The Signaling of Neuregulin-Epidermal Growth Factor Receptors and Its Impact on the Nervous System
Abstract
:1. The Family of ErbB Receptors
2. ErbB Receptor Structure
3. ErbB Receptors and Neural Development
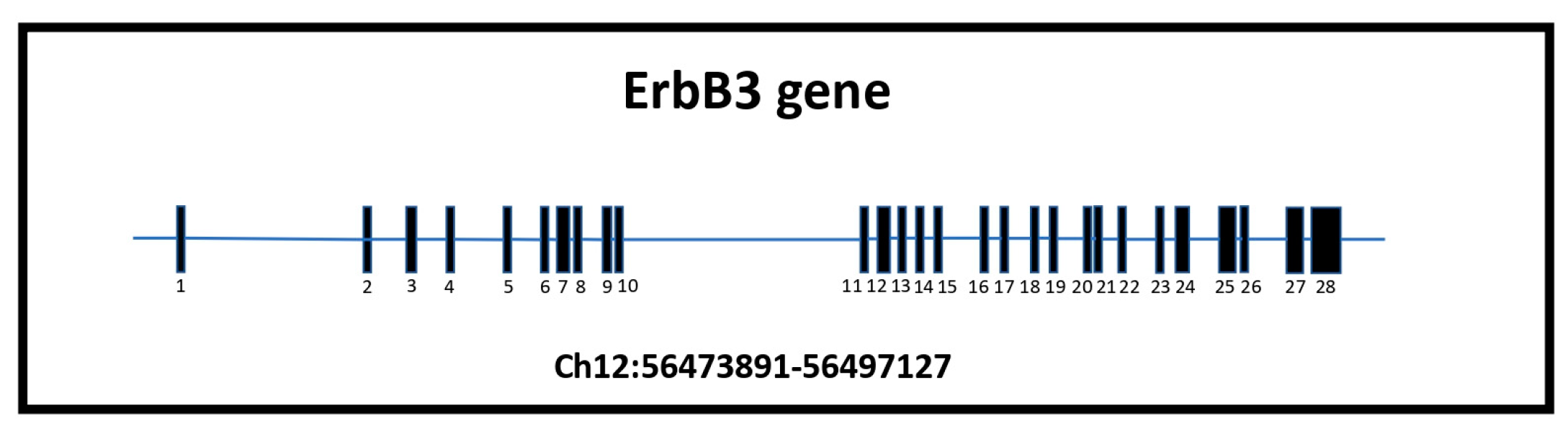
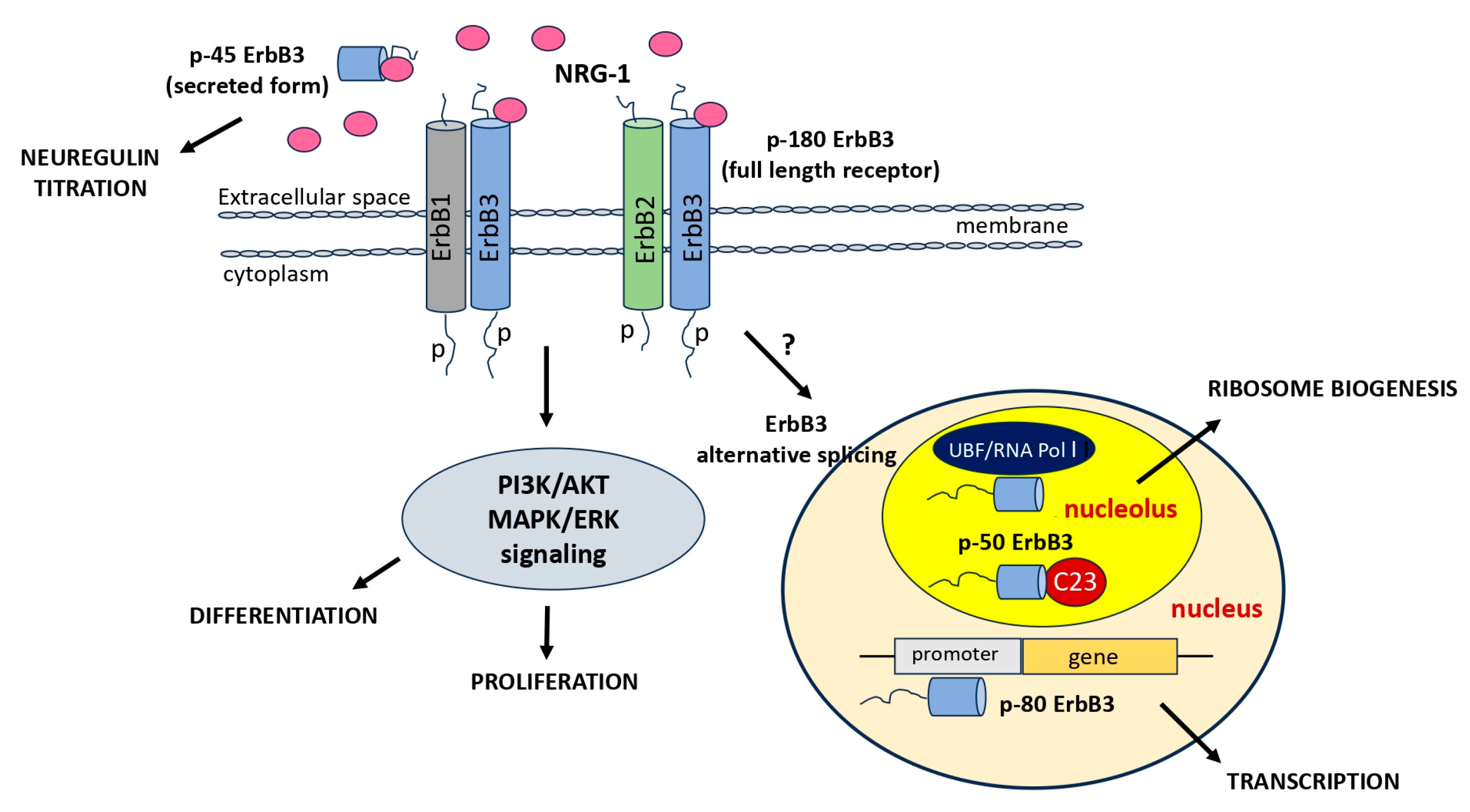
4. Focusing on ErbB3
5. Erbb3 and Glial Cells
6. ErbB3 and Myelination
7. ErbB3 and Alternative Variants
8. ErbB3 in Glioblastoma
9. ErbB3 and miRNA in Glioblastoma
10. ErbB3 Receptor and Nucleolus in Glioblastoma
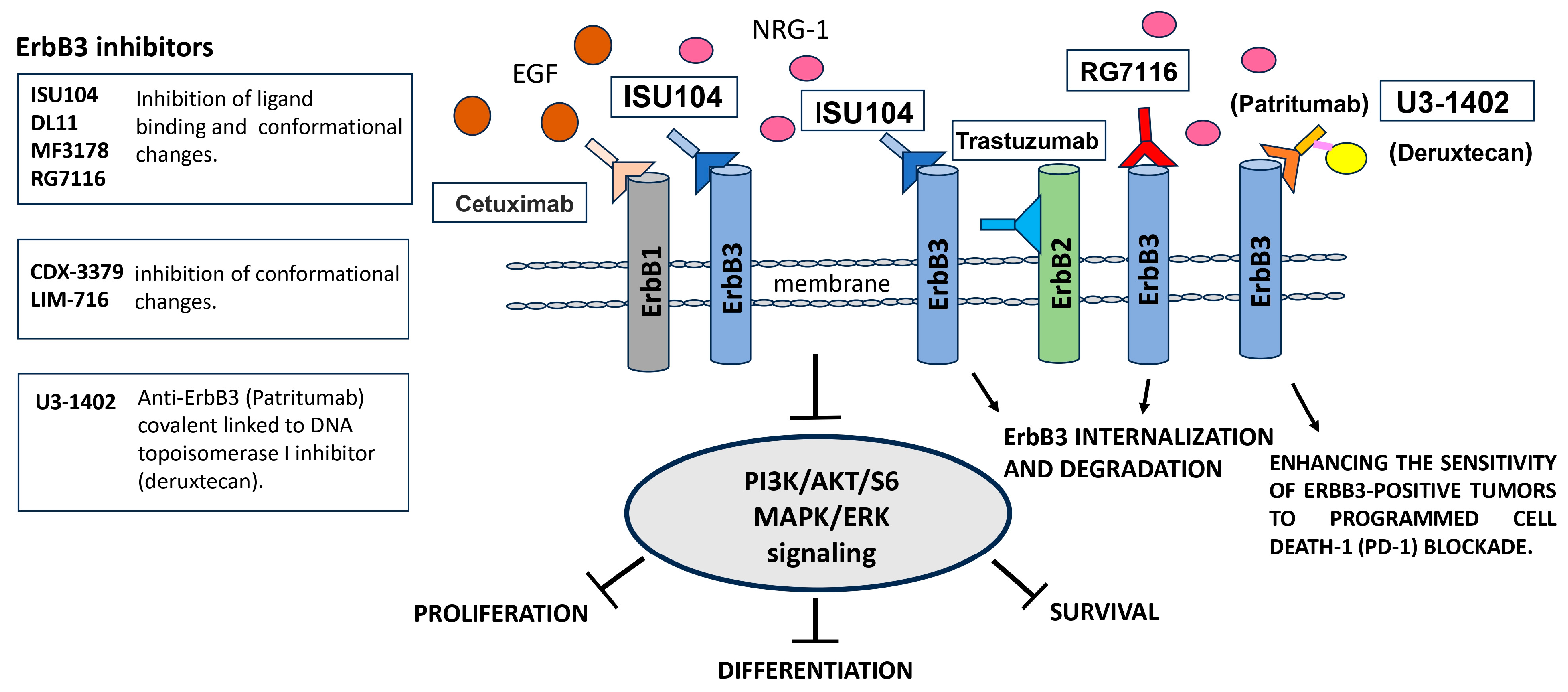
11. ErbB3 Signaling and Target Therapies
12. Discussion
13. Conclusions
14. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burgess, A.W. Regulation of Signaling from the Epidermal Growth Factor Family. J. Phys. Chem. B 2022, 126, 7475–7485. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Ferguson, K.M.; Darling, P.J.; Mohan, M.J.; Macatee, T.L.; Lemmon, M.A. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 2000, 19, 4632–4643. [Google Scholar] [CrossRef]
- Jones, J.T.; Akita, R.W.; Sliwkowski, M.X. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 1999, 447, 227–231. [Google Scholar] [CrossRef]
- Landgraf, R.; Eisenberg, D. Heregulin reverses the oligomerization of HER3. Biochemistry 2000, 39, 8503–8511. [Google Scholar] [CrossRef]
- Breuleux, M. Role of heregulin in human cancer. Cell. Mol. Life Sci. 2007, 64, 2358–2377. [Google Scholar] [CrossRef]
- Willem, M. Proteolytic processing of Neuregulin-1. Brain Res. Bull. 2016, 126 Pt 2, 178–182. [Google Scholar] [CrossRef]
- Hu, C.; Leche, C.A., 2nd; Kiyatkin, A.; Yu, Z.; Stayrook, S.E.; Ferguson, K.M.; Lemmon, M.A. Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias. Nature 2022, 602, 518–522. [Google Scholar] [CrossRef]
- Black, L.E.; Longo, J.F.; Carroll, S.L. Mechanisms of Receptor Tyrosine-Protein Kinase ErbB-3 (ERBB3) Action in Human Neoplasia. Am. J. Pathol. 2019, 189, 1898–1912. [Google Scholar] [CrossRef]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Li, D.; Yang, T.; Liu, F.; Kong, J.; Zhou, Y. PTPN1 promotes the progression of glioma by activating the MAPK/ERK and PI3K/AKT pathways and is associated with poor patient survival. Oncol. Rep. 2019, 42, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Burgess, A.W.; Cho, H.S.; Eigenbrot, C.; Ferguson, K.M.; Garrett, T.P.; Leahy, D.J.; Lemmon, M.A.; Sliwkowski, M.X.; Ward, C.W.; Yokoyama, S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 2003, 12, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.H.; Issing, W.; Miki, T.; Popescu, N.C.; Aaronson, S.A. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: Evidence for overexpression in a subset of human mammary tumors. Proc. Natl. Acad. Sci. USA 1989, 86, 9193–9197. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.; Zhang, X. The ErbB kinase domain: Structural perspectives into kinase activation and inhibition. Exp. Cell Res. 2009, 315, 649–658. [Google Scholar] [CrossRef]
- Yokoe, S.; Takahashi, M.; Asahi, M.; Lee, S.H.; Li, W.; Osumi, D.; Miyoshi, E.; Taniguchi, N. The Asn418-linked-N-glycan of ErbB3 plays a crucial role in preventing spontaneous heterodimerization and tumor promotion. Cancer Res. 2007, 67, 1935–1942. [Google Scholar] [CrossRef]
- Sotoyama, H.; Namba, H.; Tohmi, M.; Nawa, H. Schizophrenia Animal Modeling with Epidermal Growth Factor and Its Homologs: Their Connections to the Inflammatory Pathway and the Dopamine System. Biomolecules 2023, 13, 372. [Google Scholar] [CrossRef]
- Lemmetyinen, T.T.; Viitala, E.W.; Wartiovaara, L.; Kaprio, T.; Hagström, J.; Haglund, C.; Katajisto, P.; Wang, T.C.; Domènech-Moreno, E.; Ollila, S. Fibroblast derived EGF ligand neuregulin 1 induces fetal-like reprogramming of the intestinal epithelium without supporting tumorigenic growth. Dis. Models Mech. 2023, 16, dmm049692. [Google Scholar] [CrossRef]
- Ting, A.K.; Chen, Y.; Wen, L.; Yin, D.M.; Shen, C.; Tao, Y.; Liu, X.; Xiong, W.C.; Mei, L. Neuregulin 1 Promotes Excitatory Synapse Development and Function in GABAergic Interneurons. J. Neurosci. 2011, 31, 15–25. [Google Scholar] [CrossRef]
- Müller, T.; Braud, S.; Jüttner, R.; Voigt, B.C.; Paulick, K.; Sheean, M.E.; Klisch, C.; Gueneykaya, D.; Rathjen, F.G.; Geiger, J.R.; et al. Neuregulin 3 promotes excitatory synapse formation on hippocampal interneurons. EMBO J. 2018, 37, e98858. [Google Scholar] [CrossRef]
- Vullhorst, D.; Bloom, M.S.; Akella, N.; Buonanno, A. ER-PM Junctions on GABAergic Interneurons Are Organized by Neuregulin 2/VAP Interactions and Regulated by NMDA Receptors. Int. J. Mol. Sci. 2023, 24, 2908. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Nave, K.-A. Neuregulin-ERBB Signaling in the Nervous System and Neuropsychiatric Diseases. Neuron 2014, 83, 27–49. [Google Scholar] [CrossRef] [PubMed]
- Rio, C.; Rieff, H.I.; Qi, P.; Corfas, G. Neuregulin and erbB Receptors Play a Critical Role in Neuronal Migration. Neuron 1997, 19, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Namba, H.; Zheng, Y.; Nawa, H. In situ hybridization reveals developmental regulation of ErbB1-4 mRNA expression in mouse midbrain: Implication of ErbB receptors for dopaminergic neurons. Neuroscience 2009, 161, 95–110. [Google Scholar] [CrossRef]
- Iwakura, Y.; Zheng, Y.; Sibilia, M.; Abe, Y.; Piao, Y.S.; Yokomaku, D.; Wang, R.; Ishizuka, Y.; Takei, N.; Nawa, H. qualitative and quantitative re-evaluation of epidermal growth factor-ErbB1 action on developin midbrain dopaminergig action in vivo and in vitro: Target-derived neurotrophic signaling. J. Neurochem. 2011, 118, 45–56. [Google Scholar] [CrossRef]
- Makinodan, M.; Rosen, K.M.; Ito, S.; Corfas, G. A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science 2012, 337, 1357–1360. [Google Scholar] [CrossRef]
- Sathyamurthy, A.; Yin, D.M.; Barik, A.; Shen, C.; Bean, J.C.; Figueiredo, D.; She, J.X.; Xiong, W.C.; Mei, L. ERBB3-mediated regulation of Bergmann glia proliferation in cerebellar lamination. Development 2015, 142, 522–532. [Google Scholar] [CrossRef]
- Wang, H.; Liu, F.; Chen, W.; Sun, X.; Cui, W.; Dong, Z.; Zhao, K.; Zhang, H.; Li, H.; Xing, G.; et al. Genetic recovery of ErbB4 in adulthood partially restores brain functions in null mice. Proc. Natl. Acad. Sci. USA 2018, 115, 13105–13110. [Google Scholar] [CrossRef]
- Luo, B.; Liu, Z.; Lin, D.; Chen, W.; Ren, D.; Yu, Z.; Xiong, M.; Zhao, C.; Fei, E.; Li, B. ErbB4 promotes inhibitory synapse formation by cell adhesion, independent of its kinase activity. Transl. Psychiatry 2021, 11, 361. [Google Scholar] [CrossRef]
- Pitcher, J.-L.; Alexander, N.; Miranda, P.J.; Johns, T.G. ErbB4 in the brain: Focus on high grade glioma. Front. Oncol. 2022, 12, 983514. [Google Scholar] [CrossRef]
- Exposito-Alonso, D.; Osório, C.; Bernard, C.; Pascual-García, S.; Del Pino, I.; Marín, O.; Rico, B. Subcellular sorting of neuregulins controls the assembly of excitatory-inhibitory cortical circuits. eLife 2020, 9, e57000. [Google Scholar] [CrossRef] [PubMed]
- Riethmacher, D.; Sonnenberg-Riethmacher, E.; Brinkmann, V.; Yamaai, T.; Lewin, G.R.; Birchmeier, C. Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature 1997, 389, 725–730. [Google Scholar] [CrossRef]
- Hu, X.; Xiao, G.; He, L.; Zhu, Q.; Niu, X.; Li, H.; Xu, Q.; Wei, Z.; Huang, H.; Luan, Y.; et al. ErbB dysregulation impairs cognition via myelination-dependent and-independent oligodendropathy. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lyons, D.A.; Pogoda, H.M.; Voas, M.G.; Woods, I.G.; Diamond, B.; Nix, R.; Arana, N.; Jacobs, J.; Talbot, W.S. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr. Biol. 2005, 15, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Makinodan, M.; Yamauchi, T.; Tatsumi, K.; Okuda, H.; Takeda, T.; Kiuchi, K.; Sadamatsu, M.; Wanaka, A.; Kishimoto, T. Demyelination in the juvenile period, but not in adulthood, leads to long-lasting cognitive impairment and deficient social interaction in mice. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2009, 33, 978–985. [Google Scholar] [CrossRef]
- Senger, K.; Yuan, W.; Sagolla, M.; Doerr, J.; Bolon, B.; Ziai, J.; Sun, K.H.; Warming, S.; Roose-Girma, M.; Zhang, N.; et al. Embryonic lethality, and defective mammary gland development of activator-function impaired conditional knock-in Erbb3V943R mice. Adv. Genet. 2020, 2, e10036. [Google Scholar] [CrossRef]
- Limon, J.; Turc-Carel, C.; Dal Cin, P.; Rao, U.; Sandberg, A.A. Recurrent chromosome translocation in liposarcoma. Cancer Genet. Cytogenet. 1986, 22, 93–94. [Google Scholar] [CrossRef]
- Mrozek, K.; Karakousis, C.P.; Bloomfield, C.D. Chromosome 12 breakpoints are cytogenetically different in benign and malignant lipogenic tumors: Localization of breakpoints in lipoma to 12q15 and in myxoid liposarcoma to 12.q13.1. Cancer Res. 1993, 53, 1670–1675. [Google Scholar]
- Takahashi, M.; Hasegawa, Y.; Ikeda, Y.; Wada, Y.; Tajiri, M.; Ariki, S.; Takamiya, R.; Nishitani, C.; Araki, M.; Yamaguchi, Y.; et al. Suppression of heregulin β signaling by the single N-glycan deletion mutant of soluble ErbB3 protein. J. Biol. Chem. 2013, 288, 32910–32921. [Google Scholar] [CrossRef]
- Sithandam, G.; Anderson, L.M. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008, 15, 413–448. [Google Scholar] [CrossRef]
- Skinner, A.; Hurst, H.C. Transcriptional regulation of the c-erbB-3 gene in human breast carcinoma cell lines. Oncogene 1993, 8, 3393–3401. [Google Scholar]
- Zhu, C.H.; Domman, F.E. Dominant negative interference of transcription factor AP-2 causus inhibition of ErbB-3 expression and suppresses malignant cell growth. Breast Cancer Res. Treat. 2001, 71, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Kani, K.; Park, E.; Landgraf, R. The extracellular domains of ErbB3 retain high ligand binding affinty at endosome pH and in the locked conformation. Biochemistry 2005, 44, 1842–15857. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Patel, D.; Ellis, N.; Brown, S.P.; Lewandowski, J.R.; Dixon, A.M. Modulation of Transmembrane Domain Interactions in Neu Receptor Tyrosine Kinase by Membrane Fluidity and Cholesterol. J. Membr. Biol. 2019, 252, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Mendrola, J.M.; Berger, M.B.; King, M.C.; Lemmon, M.A. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002, 277, 4704–4712. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.A.; Staros, J.V. Evolutionary analysis of the ErbB receptor and ligand families. J. Mol. Evol. 2000, 50, 397–412. [Google Scholar] [CrossRef]
- Guy, P.M.; Platko, J.V.; Cantley, L.C.; Cerione, R.A.; Carraway, K.L. Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc. Natl. Acad. Sci. USA 1994, 91, 8132–8136. [Google Scholar] [CrossRef]
- Plowman, G.D.; Whitney, G.S.; Neubauer, M.G.; Green, J.M.; McDonald, V.L.; Todaro, G.J.; Shoyab, M. Molecular cloning and expression of an additional epidermal growth factor receptor-related gene. Proc. Natl. Acad. Sci. USA 1990, 87, 4905–4909. [Google Scholar] [CrossRef]
- Plowman, G.D.; Green, J.M.; Culouscou, J.M.; Carlton, G.W.; Rothwell, V.M.; Buckley, S. Heregulin induces tyrosine phosphorylation of HER4/p180erbB4. Nature 1993, 366, 473–475. [Google Scholar] [CrossRef]
- Prigent, S.A.; Gullick, W.J. Identification of c-erbB-3 binding sites for phosphaidylinositol 3′kinase and SHC using an EGF receptor/c-erbB3 chimera. EMBO J. 1994, 13, 2831–2841. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Hamburger, A.W. The use of the yeast two hybrid system to evaluate ErbB-3 interactions with SH2 domain containing proteins. Biochem. Biophys. Res. Commun. 1998, 251, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Waterman, H.; Alroy, I.; Strano, S.; Seger, R.; Yarden, Y. The C-terminus of the kinase-defective neuregulin receptor ErbB-3 confers mitogenic superiority and dictates endocytic routing. EMBO J. 1999, 18, 3348–3358. [Google Scholar] [CrossRef] [PubMed]
- Kataria, H.; Alizadeh, A.; Karimi-Abdolrezaee, S. Neuregulin-1/ErbB network: An emerging modulator of nervous system injury and repair. Prog. Neurobiol. 2019, 180, 101643. [Google Scholar] [CrossRef] [PubMed]
- Goldowitz, D.; Hamre, K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998, 21, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Manni, E.; Petrosini, L. A century of cerebellar somatotopy: A debated representation. Nat. Rev. Neurosci. 2004, 5, 241–249. [Google Scholar] [CrossRef]
- Bignami, A.; Dahl, D. Differentiation of astrocytes in the cerebellar cortex and the pyramidal tracts of the newborn rat. An immunofluorescence study with antibodies to a protein specific to astrocytes. Brain Res. 1973, 49, 393–402. [Google Scholar] [CrossRef]
- Yamada, K.; Watanabe, M. Cytodifferentiation of Bergmann glia and its relationship with Purkinje cells. Anat. Sci. Int. 2002, 77, 94–108. [Google Scholar] [CrossRef]
- Ledonne, A.; Mercuri, N.B. On the Modulatory Roles of Neuregulins/ErbB Signaling on Synaptic Plasticity. Int. J. Mol. Sci. 2019, 21, 275. [Google Scholar] [CrossRef]
- Busfield, S.J.; Michnick, D.A.; Chickering, T.W.; Revett, T.L.; Ma, J.; Woolf, E.A.; Comrack, C.A.; Dussault, B.J.; Woolf, J.; Goodearl, A.D.; et al. Characterization of a neuregulin-related gene, Don-1, that is highly expressed in restricted regions of the cerebellum and hippocampus. Mol. Cell. Biol. 1997, 17, 4007–4014. [Google Scholar] [CrossRef]
- Carraway, K.L., III; Weber, J.L.; Unger, M.J.; Ledesma, J.; Yu, N.; Gassmann, M.; Lai, C. Neuregulin-2, a new ligand of ErbB3/ErbB4-receptor tyrosine kinases. Nature 1997, 387, 512–516. [Google Scholar] [CrossRef]
- Jüttner, R.; Rathjen, F.G. Molecular analysis of axonal target specificity and synapse formation. Cell. Mol. Life Sci. 2005, 62, 2811–2827. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, Y.; Ishiguro, H.; Tokita, Y.; Oohira, A.; Ohmoto, H.; Higashiyama, S. Neuroglycan C, a novel member of the neuregulin family. Biochem. Biophys. Res. Commun. 2004, 321, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Fox, I.J.; Kornblum, H.I. Developmental Profiling of ErbB Receptors in the Murine Central Nervous System: Implications for Functional Interactions. J. Neurosci. Res. 2005, 79, 584–597. [Google Scholar] [CrossRef]
- Francoeur, J.R.; Richardson, P.M.; Dunn, R.J.; Carbonetto, S. Distribution of Erb-B2, Erb-B3, and Erb-B4 in the Developing Avian Nervous System. J. Neurosci. Res. 1995, 41, 836–845. [Google Scholar] [CrossRef]
- Thompson, M.; Lauderdale, S.; Webster, M.J.; Chong, V.Z.; McClintock, B.; Saunders, R.; Weickert, C.S. Widespread expression of ErbB2, ErbB3 and ErbB4 in non-human primate brain. Brain Res. 2007, 1139, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Grider, M.H.; Belcea, C.Q.; Covington, B.P.; Reddy, V.; Sharma, S. Neuroanatomy, Nodes of Ranvier; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Snaidero, N.; Möbius, W.; Czopka, T.; Hekking, L.H.; Mathisen, C.; Verkleij, D.; Goebbels, S.; Edgar, J.; Merkler, D.; Lyons, D.A.; et al. Myelin membrane wrapping of CNS axons by PI (3,4,5) P3-dependent polarized growth at the inner tongue. Cell 2014, 156, 277–290. [Google Scholar] [CrossRef]
- Emery, B. Transcriptional and post-transcriptional control of CNS myelination. Curr. Opin. Neurobiol. 2010, 20, 601–607. [Google Scholar] [CrossRef]
- Emery, B. Regulation of oligodendrocyte differentiation and myelination. Science 2010, 330, 779–782. [Google Scholar] [CrossRef]
- Nave, K.A. Myelination, and support of axonal integrity by glia. Nature 2010, 468, 244–252. [Google Scholar] [CrossRef]
- Lubetzki, C.; Sol-Foulon, N.; Desmazières, A. Nodes of Ranvier during development and repair in the CNS. Nat. Rev. Neurol. 2020, 16, 426–439. [Google Scholar] [CrossRef]
- Fünfschilling, U.; Supplie, L.M.; Mahad, D.; Boretius, S.; Saab, A.S.; Edgar, J.; Brinkmann, B.G.; Kassmann, C.M.; Tzvetanova, I.D.; Möbius, W.; et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 2012, 485, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Ronzano, R.; Roux, T.; Thetiot, M.; Aigrot, M.S.; Richard, L.; Lejeune, F.X.; Mazuir, E.; Vallat, J.M.; Lubetzki, C.; Desmazières, A. Microglia-neuron interaction at nodes of Ranvier depends on neuronal activity through potassium release and contributes to remyelination. Nat. Commun. 2021, 12, 5219. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K.D.; Partlow, L.M. Preparation of pure neuronal and non-neuronal cultures from embryonic chick sympathetic ganglia: A new method based on both differential cell adhesiveness and the formation of homotypic neuronal aggregates. Brain Res. 1976, 114, 391–414. [Google Scholar] [CrossRef] [PubMed]
- Salzer, J.L.; Williams, A.K.; Glaser, L.; Bunge, R.P. Studies of Schwann cell proliferation. II. Characterization of the stimulation and specificity of the response to a neurite membrane fraction. J. Cell Biol. 1980, 84, 753–766. [Google Scholar] [CrossRef]
- Wood, P.M.; Bunge, R.P. Evidence that sensory axons are mitogenic for Schwann cells. Nature 1975, 256, 662–664. [Google Scholar] [CrossRef]
- Lemke, G.E.; Brockes, J.P. Identification and purification of glial growth factor. J. Neurosci. 1984, 4, 75–83. [Google Scholar] [CrossRef]
- Meyer, D.; Birchmeier, C. Multiple essential functions of neuregulin in development. Nature 1995, 378, 386–390. [Google Scholar] [CrossRef]
- Kim, K.; Lee, D. ERBB3-dependent AKT and ERK pathways are essential for atrioventricular cushion development in mouse embryos. PLoS ONE 2021, 16, e0259426. [Google Scholar] [CrossRef]
- Wolpowitz, D.; Mason, T.B.; Dietrich, P.; Mendelsohn, M.; Talmage, D.A.; Role, L.W. Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron 2000, 25, 79–91. [Google Scholar] [CrossRef]
- McCarthy, K.D.; Partlow, L.M. Neuronal stimulation of (3H) thymidine incorporation by primary cultures of highly purified non-neuronal cells. Brain Res. 1976, 114, 415–426. [Google Scholar] [CrossRef]
- Salzer, J.L.; Bunge, R.P. Studies of Schwann cell proliferation. I. An analysis in tissue culture of proliferation during development, Wallerian degeneration, and direct injury. J. Cell Biol. 1980, 84, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Honjo, Y.; Kniss, J.; Eisen, J.S. Neuregulin-mediated ErbB3 signaling is required for formation of zebrafish dorsal root ganglion neurons. Development 2008, 135, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Garratt, A.N.; Voiculescu, O.; Topilko, P.; Charnay, P.; Birchmeier, C. A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J. Cell Biol. 2000, 148, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Akmentin, W.; Du, C.; Role, L.W.; Talmage, D.A. Axonal Type III Nrg1 Controls Glutamate Synapse Formation and GluA2 Trafficking in Hippocampal-Accumbens Connections. eNeuro 2017, 4, ENEURO.0232-16.2017. [Google Scholar] [CrossRef]
- Andrique, L.; Fauvin, D.; El Maassarani, M.; Colasson, H.; Vannier, B.; Séité, P. ErbB3 80 kDa, a nuclear variant of the ErbB3 receptor, binds to the Cyclin D1 promoter to activate cell proliferation but is negatively controlled by p14ARF. Cell. Signal. 2012, 24, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Adilakshmi, T.; Ness-Myers, J.; Madrid-Aliste, C.; Fiser, A.; Tapinos, N. A nuclear variant of ErbB3 receptor tyrosine kinase regulates ezrin distribution and Schwann cell myelination. J. Neurosci. 2011, 31, 5106–5119. [Google Scholar] [CrossRef] [PubMed]
- Jathal, M.K.; Siddiqui, S.; Vasilatis, D.M.; Durbin Johnson, B.P.; Drake, C.; Mooso, B.A.; D’Abronzo, L.S.; Batra, N.; Mudryj, M.; Ghosh, P.M. Androgen receptor transcriptional activity is required for heregulin-1β-mediated nuclear localization of the HER3/ErbB3 receptor tyrosine kinase. J. Biol. Chem. 2023, 299, 104973. [Google Scholar] [CrossRef]
- Melendez-Vasquez, C.V.; Rios, J.C.; Zanazzi, G.; Lambert, S.; Bretscher, A.; Salzer, J.L. Nodes of Ranvier form in association with ezrin-radixin-moesin (ERM)-positive Schwann cell processes. Proc. Natl. Acad. Sci. USA 2001, 98, 1235–1240. [Google Scholar] [CrossRef]
- Rasband, M.N.; Peles, E. Mechanisms of node of Ranvier assembly. Nat. Rev. Neurosci. 2021, 22, 7–20. [Google Scholar] [CrossRef]
- Katoh, M.; Yazaki, Y.; Sugimura, T.; Terada, M. c-erbB3 gene encodes secreted as well transmembrane receptor tyrosine kinase. Biochem. Biophys. Res. Commun. 1993, 192, 1189–1197. [Google Scholar] [CrossRef]
- Srinivasan, R.; Leverton, K.E.; Sheldon, H.; Hurst, H.C.; Sarraf, C.; Gullick, W.J. Intracellular expression of the truncated extracellular domain of c-erbB3/HER3. Cell. Signal. 2001, 13, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Maihle, N.J. Isolation and characterization of four alternate c-erbB3 transcripts expressed in ovarian carcinoma-derived cell lines and normal human tissues. Oncogene 1998, 16, 3243–3252. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Akita, R.W.; Sliwkowski, M.X.; Maihle, N.J. A naturally occurring secreted human ErbB3 receptor isoform inhibits heregulin-stimulated activation of ErbB2, ErbB3 and ErbB4. Cancer Res. 2001, 61, 4467–4473. [Google Scholar]
- Tagliaferro, M.; Rosa, P.; Bellenchi, G.C.; Bastianelli, D.; Trotta, R.; Tito, C.; Fazi, F.; Calogero, A.; Ponti, D. Nucleolar localization of the ErbB3 receptor as a new target in glioblastoma. BMC Mol. Cell Biol. 2022, 23, 13. [Google Scholar] [CrossRef] [PubMed]
- Fuentealba, L.C.; Obernier, K.; Alvarez-Buylla, A. Adult neural stem cells bridge their niche. Cell Stem Cell 2012, 10, 698–708. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncology 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109, Erratum in Acta Neuropathol. 2007, 114, 547. [Google Scholar] [CrossRef]
- Bai, J.; Varghese, J.; Jain, R. Adult Glioma WHO Classification Update, Genomics, and Imaging: What the Radiologists Need to Know. Top. Magn. Reson. Imaging 2020, 29, 71–82. [Google Scholar] [CrossRef]
- Riquelme, P.A.; Drapeau, E.; Doetsch, F. Brain micro-ecologies: Neural stem cell niches in the adult mammalian brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 123–137. [Google Scholar] [CrossRef]
- Fisher, J.L.; Schwartzbaum, J.A.; Wrensch, M.; Wiemels, J.L. Epidemiology of brain tumors. Neurol. Clin. 2007, 25, 867–890. [Google Scholar] [CrossRef]
- Linos, E.; Raine, T.; Alonso, A.; Michaud, D. Atopy and risk of brain tumors: A meta-analysis. J. Natl. Cancer Inst. 2007, 99, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Wrensch, M.; Wiencke, J.K.; Wiemels, J.; Miike, R.; Patoka, J.; Moghadassi, M.; McMillan, A.; Kelsey, K.T.; Aldape, K.; Lamborn, K.R.; et al. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006, 66, 4531–4541. [Google Scholar] [CrossRef] [PubMed]
- Wirsching, H.G.; Galanis, E.; Weller, M. Glioblastoma. Handb. Clin. Neurol. 2016, 134, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477, Erratum in Cell 2014, 157, 753. [Google Scholar] [CrossRef]
- Farrell, C.J.; Plotkin, S.R. Genetic causes of brain tumors: Neurofibromatosis, tuberous sclerosis, von Hippel-Lindau, and other syndromes. Neurol. Clin. 2007, 25, 925–946. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Ransom, D.T.; Ritland, S.R.; Kimmel, D.W.; Moertel, C.A.; Dahl, R.J.; Scheithauer, B.W.; Kelly, P.J.; Jenkins, R.B. Cytogenetic and loss of heterozygosity studies in ependymomas, pilocytic astrocytomas, and oligodendrogliomas. Genes Chromosomes Cancer 1992, 5, 348–356. [Google Scholar] [CrossRef]
- Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Glioblastoma: From molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014, 9, 1–25. [Google Scholar] [CrossRef]
- Kleihues, P.; Ohgaki, H. Primary and secondary glioblastomas: From concept to clinical diagnosis. Neuro-Oncology 1999, 1, 44–51. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Chen, X.; Zhang, S.; Shi, H.; Ye, Y.; Shi, H.; Zou, Z.; Li, P.; Guo, Q.; Ma, L.; et al. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 2021, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Furnari, F.B.; Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat. Rev. Cancer 2015, 15, 302–310. [Google Scholar] [CrossRef]
- Andersson, U.; Guo, D.; Malmer, B.; Bergenheim, A.T.; Brännström, T.; Hedman, H.; Henriksson, R. Epidermal growth factor receptor family (EGFR, ErbB2-4) in gliomas and meningiomas. Acta Neuropathol. 2004, 108, 135–142. [Google Scholar] [CrossRef]
- Stommel, J.M.; Kimmelman, A.C.; Ying, H.; Nabioullin, R.; Ponugoti, A.H.; Wiedemeyer, R.; Stegh, A.H.; Bradner, J.E.; Ligon, K.L.; Brennan, C.; et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 2007, 318, 287–290. [Google Scholar] [CrossRef]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genßler, S.; Schönfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2015, 108, djv375. [Google Scholar] [CrossRef]
- De Bacco, F.; Orzan, F.; Erriquez, J.; Casanova, E.; Barault, L.; Albano, R.; D’Ambrosio, A.; Bigatto, V.; Reato, G.; Patanè, M.; et al. ERBB3 overexpression due to miR-205 inactivation confers sensitivity to FGF, metabolic activation, and liability to ERBB3 targeting in glioblastoma. Cell Rep. 2021, 36, 109455. [Google Scholar] [CrossRef]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023. [Google Scholar] [CrossRef]
- Lukiw, W.J. MicroRNA (miRNA) Complexity in Alzheimer’s Disease (AD). Biology 2023, 12, 788. [Google Scholar] [CrossRef]
- Ishida, T.; Ueyama, T.; Ihara, D.; Harada, Y.; Nakagawa, S.; Saito, K.; Nakao, S.; Kawamura, T. c-Myc/microRNA-17-92 Axis Phase-Dependently Regulates PTEN and p21 Expression via ceRNA during Reprogramming to Mouse Pluripotent Stem Cells. Biomedicines 2023, 11, 1737. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Mafi, A.; Rahmati, A.; Babaei Aghdam, Z.; Salami, R.; Salami, M.; Vakili, O.; Aghadavod, E. Recent insights into the microRNA-dependent modulation of gliomas from pathogenesis to diagnosis and treatment. Cell. Mol. Biol. Lett. 2022, 27, 65. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Asadi-Moghaddam, K.; Chiocca, E.A.; Lawler, S.E. Potential role of miRNAs and their inhibitors in glioma treatment. Expert Rev. Anticancer Ther. 2010, 10, 1753–1762. [Google Scholar] [CrossRef]
- Guessous, F.; Zhang, Y.; Kofman, A.; Catania, A.; Li, Y.; Schiff, D.; Purow, B.; Abounader, R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 2010, 9, 1031–1036. [Google Scholar] [CrossRef]
- Yuan, M.; Da Silva, A.C.A.L.; Arnold, A.; Okeke, L.; Ames, H.; Correa-Cerro, L.S.; Vizcaino, M.A.; Ho, C.Y.; Eberhart, C.G.; Rodriguez, F.J. MicroRNA (miR) 125b regulates cell growth and invasion in pediatric low grade glioma. Sci. Rep. 2018, 8, 12506. [Google Scholar] [CrossRef]
- Wang, D.D.; Ma, L.; Wong, M.P.; Lee, V.H.; Yan, H. Contribution of EGFR and ErbB-3 Heterodimerization to the EGFR Mutation-Induced Gefitinib and Erlotinib-Resistance in Non-Small-Cell Lung Carcinoma Treatments. PLoS ONE 2015, 10, e0128360. [Google Scholar] [CrossRef]
- Koganemaru, S.; Kuboki, Y.; Koga, Y.; Kojima, T.; Yamauchi, M.; Maeda, N.; Kagari, T.; Hirotani, K.; Yasunaga, M.; Matsumura, Y.; et al. U3-1402, a Novel HER3-Targeting Antibody-Drug Conjugate, for the Treatment of Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 2043–2050. [Google Scholar] [CrossRef]
- Elhamamsy, A.R.; Metge, B.J.; Alsheikh, H.A.; Shevde, L.A.; Samant, R.S. Ribosome Biogenesis: A Central Player in Cancer Metastasis and Therapeutic Resistance. Cancer Res. 2022, 82, 2344–2353. [Google Scholar] [CrossRef]
- Zhai, W.; Comai, L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 2000, 20, 5930–5938. [Google Scholar] [CrossRef] [PubMed]
- Voit, R.; Schäfer, K.; Grummt, I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol. 1997, 17, 4230–4237. [Google Scholar] [CrossRef] [PubMed]
- Ponti, D.; Bellenchi, G.C.; Puca, R.; Bastianelli, D.; Maroder, M.; Ragona, G.; Roussel, P.; Thiry, M.; Mercola, D.; Calogero, A. The transcription factor EGR1 localizes to the nucleolus and is linked to suppression of ribosomal precursor synthesis. PLoS ONE 2014, 9, e96037. [Google Scholar] [CrossRef] [PubMed]
- Ponti, D.; Bastianelli, D.; Rosa, P.; Pacini, L.; Ibrahim, M.; Rendina, E.A.; Ragona, G.; Calogero, A. The expression of B23 and EGR1 proteins is functionally linked in tumor cells under stress conditions. BMC Cell Biol. 2015, 16, 27. [Google Scholar] [CrossRef]
- Madera, S.; Izzo, F.; Chervo, M.F.; Dupont, A.; Chiauzzi, V.A.; Bruni, S.; Petrillo, E.; Merin, S.S.; De Martino, M.; Montero, D.; et al. Halting ErbB-2 isoforms retrograde transport to the nucleus as a new theragnostic approach for triple-negative breast cancer. Cell Death Dis. 2022, 13, 447. [Google Scholar] [CrossRef]
- Wang, Y.N.; Yamaguchi, H.; Hsu, J.M.; Hung, M.C. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 2010, 29, 3997–4006. [Google Scholar] [CrossRef]
- Sundvall, M.; Peri, L.; Määttä, J.A.; Tvorogov, D.; Paatero, I.; Savisalo, M.; Silvennoinen, O.; Yarden, Y.; Elenius, K. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene 2007, 26, 6905–6914. [Google Scholar] [CrossRef]
- Farin, K.; Di Segni, A.; Mor, A.; Pinkas-Kramarski, R. Structure-function analysis of nucleolin and ErbB receptors interactions. PLoS ONE 2009, 4, e6128. [Google Scholar] [CrossRef]
- Wang, M.; Trim, C.M.; Gullick, W.J. Localisation of Neuregulin 1-β3 to different sub-nuclear structures alters gene expression. Exp. Cell Res. 2011, 317, 423–432. [Google Scholar] [CrossRef]
- Kumagai, S.; Koyama, S.; Nishikawa, H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 2021, 21, 181–197. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- De Bacco, F.; Boccaccio, C. ERBB3 as a therapeutic target in glioblastoma: Overexpression can make the difference. Mol. Cell. Oncol. 2021, 8, 1990677. [Google Scholar] [CrossRef] [PubMed]
- Ocana, A.; Vera-Badillo, F.; Seruga, B.; Templeton, A.; Pandiella, A.; Amir, E. HER3 overexpression and survival in solid tumors: A meta-analysis. J. Natl. Cancer Inst. 2013, 105, 266–273, Erratum in J. Natl. Cancer Inst. 2013, 105, 944. [Google Scholar] [CrossRef] [PubMed]
- Haratani, K.; Yonesaka, K.; Takamura, S.; Maenishi, O.; Kato, R.; Takegawa, N.; Kawakami, H.; Tanaka, K.; Hayashi, H.; Takeda, M.; et al. U3-1402 sensitizes HER3-expressing tumors to PD-1 blockade by immune activation. J. Clin. Investig. 2020, 130, 374–388. [Google Scholar] [CrossRef]
- Hong, M.; Yoo, Y.; Kim, M.; Kim, J.Y.; Cha, J.S.; Choi, M.K.; Kim, U.; Kim, K.; Sohn, Y.; Bae, D.; et al. A Novel Therapeutic Anti-ErbB3, ISU104 Exhibits Potent Antitumorigenic Activity by Inhibiting Ligand Binding and ErbB3 Heterodimerization. Mol. Cancer Ther. 2021, 20, 1142–1152. [Google Scholar] [CrossRef]
- Cejalvo, J.M.; Jacob, W.; Fleitas Kanonnikoff, T.; Felip, E.; Navarro Mendivil, A.; Martinez Garcia, M.; Taus Garcia, A.; Leighl, N.; Lassen, U.; Mau-Soerensen, M.; et al. A phase Ib/II study of HER3-targeting lumretuzumab in combination with carboplatin and paclitaxel as first-line treatment in patients with advanced or metastatic squamous non-small cell lung cancer. ESMO Open 2019, 4, e000532. [Google Scholar] [CrossRef]
- Mirschberger, C.; Schiller, C.B.; Schräml, M.; Dimoudis, N.; Friess, T.; Gerdes, C.A.; Reiff, U.; Lifke, V.; Hoelzlwimmer, G.; Kolm, I.; et al. RG7116, a therapeutic antibody that binds the inactive HER3 receptor and is optimized for immune effector activation. Cancer Res. 2013, 73, 5183–5194. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Koyama, K.; Kamai, Y.; Hirotani, K.; Ogitani, Y.; Zembutsu, A.; Abe, M.; Kaneda, Y.; Maeda, N.; Shiose, Y.; et al. A Novel HER3-Targeting Antibody-Drug Conjugate, U3-1402, Exhibits Potent Therapeutic Efficacy through the Delivery of Cytotoxic Payload by Efficient Internalization. Clin. Cancer Res. 2019, 25, 7151–7161. [Google Scholar] [CrossRef]
- Gandullo-Sánchez, L.; Ocaña, A.; Pandiella, A. HER3 in cancer: From the bench to the bedside. J. Exp. Clin. Cancer Res. 2022, 41, 310. [Google Scholar] [CrossRef]
- Pan, P.C.; Magge, R.S. Mechanisms of EGFR Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 8471. [Google Scholar] [CrossRef]
- Hafeez, U.; Parslow, A.C.; Gan, H.K.; Scott, A.M. New insights into ErbB3 function and therapeutic targeting in cancer. Expert Rev. Anticancer Ther. 2020, 20, 1057–1074. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tagliaferro, M.; Ponti, D. The Signaling of Neuregulin-Epidermal Growth Factor Receptors and Its Impact on the Nervous System. Neuroglia 2023, 4, 253-274. https://doi.org/10.3390/neuroglia4040018
Tagliaferro M, Ponti D. The Signaling of Neuregulin-Epidermal Growth Factor Receptors and Its Impact on the Nervous System. Neuroglia. 2023; 4(4):253-274. https://doi.org/10.3390/neuroglia4040018
Chicago/Turabian StyleTagliaferro, Marzia, and Donatella Ponti. 2023. "The Signaling of Neuregulin-Epidermal Growth Factor Receptors and Its Impact on the Nervous System" Neuroglia 4, no. 4: 253-274. https://doi.org/10.3390/neuroglia4040018



