The Comparison of Mutational Progression in SARS-CoV-2: A Short Updated Overview
Abstract
:Highlights
- Understanding of mutations provides structural insight into SARS-CoV-2 variants;
- Comparison of mutations narrates the common and different mutations in the variants of concern (VOCs);
- The SD1-SD2 domain and S2 subunit provides an opportunistic spot for future virulence and vaccine development studies on SARS-CoV-2.
- The SARS-CoV-2 epidemic has paralyzed the healthcare system, shifting the focus of science. It is crucial to comprehend the mutations documented in the Beta (B.1.351), Gamma (P.1), Delta (B.1.612.2), and Omicron (B.1.1.529) variants, which will help us comprehend any future concerns related to COVID 19 and could pave the way for defense strategies against any future variants.
Abstract
1. Introduction
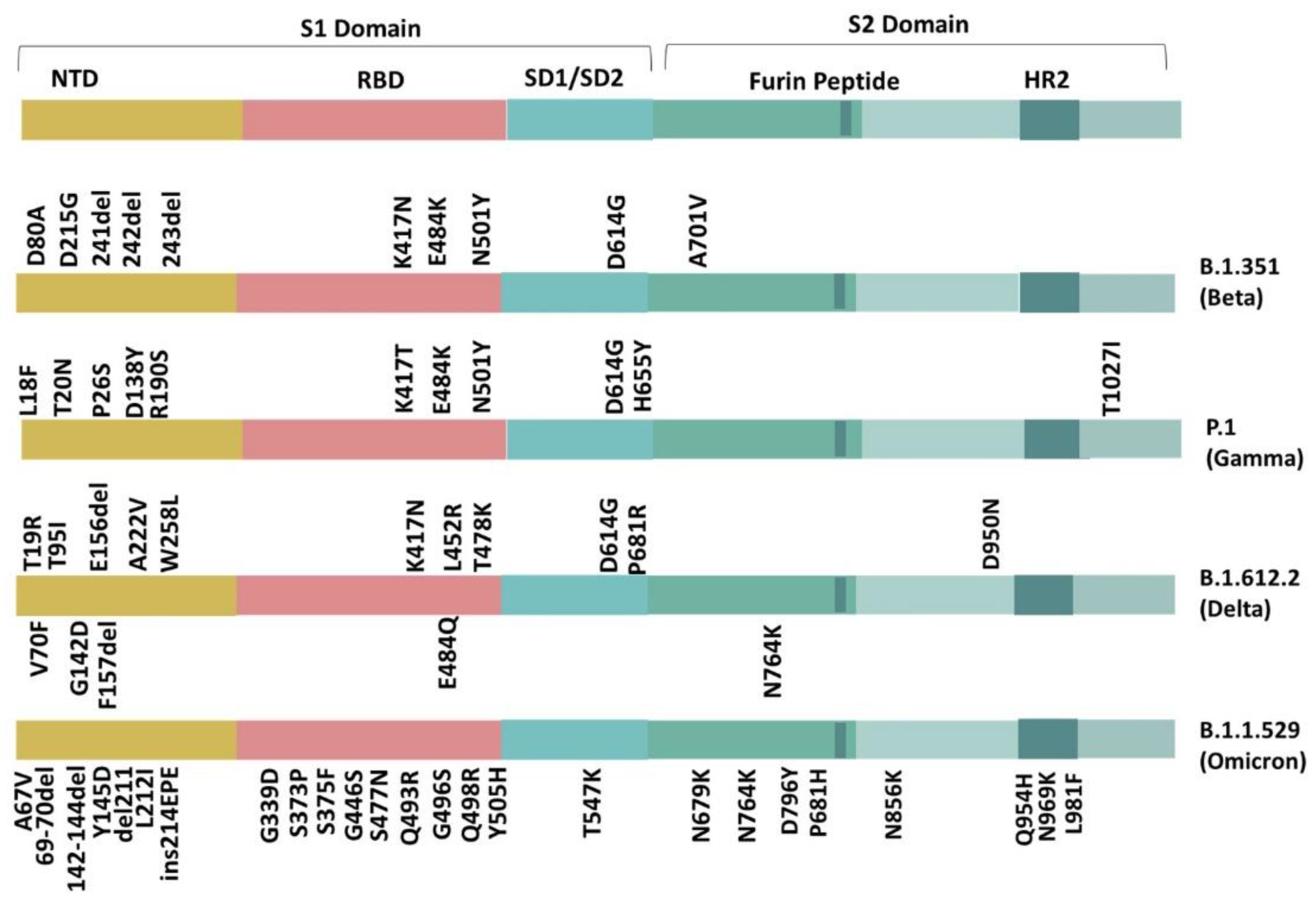
2. Variants of SARS-CoV-2
3. Identified Mutations according to Their Location in the Beta, Gamma, Delta, and Omicron Variant
3.1. S1 Domain (N-terminal Domain (NTD))
3.2. Receptor Binding Domain (RBD)
3.3. SD1-SD2 Region
3.4. S2 Domain
| Mutation | Beta | Gamma | Delta | Omicron | Domain | Location | Mutation Effect |
|---|---|---|---|---|---|---|---|
| L18F | . | ✔ | . | . | S1 Domain | NTD | It hinders the binding of antibodies and increases the virulence [83]. |
| T19R | . | . | ✔ | . | S1 Domain | NTD | It increases the binding of the viral particle with the ACE2 protein and increases its evasiveness against antibodies [28]. |
| T20N | . | ✔ | . | . | S1 Domain | NTD | It brings about the glycan shielding of supersites through glycosylation, which alters the binding capacity of the antibodies [19]. |
| P26S | . | ✔ | . | . | S1 Domain | NTD | It brings strong immune evasion [20]. |
| A67V | . | . | . | ✔ | S1 Domain | NTD | It induces conformational changeswithin the N2 loop and enhances viral infectivity [37]. |
| H69-V70 del | . | . | . | ✔ | S1 Domain | NTD | It increases viral transmission and evasiveness [33,35,84]. |
| V70F | . | . | ✔ | . | S1 Domain | NTD | It tends to change the secondary structure of the protein [31]. |
| D80A | ✔ | . | . | . | S1 Domain | NTD | - |
| T95I | . | . | ✔ | . | S1 Domain | NTD | Increases virulence [29]. |
| D138Y | . | ✔ | . | . | S1 Domain | NTD | - |
| del142-144 | . | . | . | ✔ | S1 Domain | NTD | It enhances infectivity, evasiveness, and shifts an N3 central loop [33,34,35]. |
| G142D | . | . | ✔ | . | S1 Domain | NTD | It produces structural changes [29]. |
| Y145D | . | . | . | ✔ | S1 Domain | NTD | It increases viral evasiveness and reduces neutralization antibodies’ effectivity [25,38]. |
| E156- and F157- | . | . | ✔ | . | S1 Domain | NTD | - |
| R158G | . | . | ✔ | . | S1 Domain | NTD | Reduces the neutralization of monoclonal antibodies [29,30]. |
| R190S | . | ✔ | . | . | S1 Domain | NTD | Enhances viral immunity [20,21]. |
| del211 | . | . | . | ✔ | S1 Domain | NTD | It induces configurational changes [37]. |
| L212I | . | . | . | ✔ | S1 Domain | NTD | It reduces the binding affinity of some monoclonal antibodies [26]. |
| D215G | ✔ | . | . | . | S1 Domain | NTD | - |
| A222V | . | . | ✔ | . | S1 Domain | NTD | It increases the opening of RBD, slightly increasing ACE2 affinity, but not evasive capabilities [22,23,24].. |
| 241/242/243del | ✔ | . | . | . | S1 Domain | NTD | - |
| ins214EPE | . | . | . | ✔ | S1 Domain | NTD | It induces a structural change in the loop in the 209-216 residue and increases viral evasiveness [37]. |
| W258L | . | . | ✔ | . | S1 Domain | NTD | It skews the R246 side chain [36]. |
| G339D | . | . | . | ✔ | S1 Domain | RBD | It induces a slight increase in the binding affinity to ACE2 and improves virus evasion against some neutralizing antibodies [46]. |
| S371L | . | . | . | ✔ | S1 Domain | RBD | It instigates a conformational change across antigenic sites, inducing antibody resistance and increasing ACE2 binding affinity [45,46,47,48,49] |
| S373P | . | . | . | ✔ | S1 Domain | RBD | It induces a conformational change in α2 and β2 loop and antigenic site. It also causes an increase in the binding affinity to ACE2 [45,46,47,48]. |
| S375F | . | . | . | ✔ | S1 Domain | RBD | It induces a change in antigenic site and improves viral evasion [46]. |
| K417T | . | ✔ | . | . | S1 Domain | RBD | It increases immune escape [40,44]. |
| K417N | ✔ | . | ✔ | . | S1 Domain | RBD | It enhances virulence and antibody immunization [41,42,47]. |
| N440K | . | . | . | ✔ | S1 Domain | RBM | It increases viral evasiveness against antibodies [46,49]. |
| G446S | . | . | . | ✔ | S1 Domain | RBM | It induces steric hindrance and reduces antibody activity against the virus [23,49,62]. |
| L452R | . | . | ✔ | . | S1 Domain | RBM | It leads to a more stable S subunit and a higher affinity for the ACE2 receptor [43,85]. |
| S477N | . | . | . | ✔ | S1 Domain | RBM | It alters the antigenic traits of the virus, improving the virus’s evasion against antibodies [37]. |
| T478K | . | . | ✔ | . | S1 Domain | RBM | It engenders increased steric hindrance [61]. |
| E484Q | . | . | ✔ | . | S1 Domain | RBM | Increases virulence and virus stability due to disrupted hydrogen bonds [43,51]. |
| E484K | ✔ | ✔ | . | . | S1 Domain | RBM | Enhances antibody evasion [55]. |
| E484A | . | . | . | . | S1 Domain | RBM | It weakens the interactions with ACE2 but increases viral evasion against antibodies [37,46]. |
| Q493R | . | . | . | ✔ | S1 Domain | RBM | It contributes to increased ACE2 binding affinity and immune evasion by causing steric hindrance [23,37,48,49]. |
| G496S | . | . | . | ✔ | S1 Domain | RBM | It strengthens viral affinity to ACE2 and improves viral evasiveness against antibodies [37,46,62]. |
| Q498R | . | . | . | . | S1 Domain | RBM | It improves viral affinity to ACE2 [37,48,86]. |
| N501Y | ✔ | ✔ | . | . | S1 Domain | RBM | It increases viral affinity with ACE2 and increases viral entry into the host [57,59]. |
| Y505H | . | . | . | ✔ | S1 Domain | RBM | It improves virus evasiveness against casirivimab, a monoclonal antibody [23,46]. |
| T547K | . | . | . | ✔ | S1 Domain | SD1-SD2 | It improves interactions between S1 and S2, destabilizes spike protein structure, and decreases spike protein flexibility [66,67,68]. |
| D614G | ✔ | ✔ | ✔ | ✔ | S1 Domain | SD1-SD2 | It increases virulence but also makes it increasingly susceptible to neutralization [30,70]. |
| H655Y | . | ✔ | . | ✔ | S1 Domain | SD1-SD2 | Has a positive effect on furin cleavage and enhances the transmission of the virus [44]. |
| N679K | . | . | . | ✔ | S2 Domain | Near Furin Cleavage Site | It makes the furin cleavage site more polybasic and increases its cleavability [76]. It also encourages glycosylation at the cleavage site, discouraging syncytia formation [75]. |
| P681R | . | . | ✔ | . | S2 Domain | Near Furin Cleavage Site | It allows a more efficient viral entry [71]. |
| P681H | . | . | . | ✔ | S2 Domain | Near Furin Cleavage Site | It enhances spike cleavage, viral infection, and offers adaptive immunity but does not significantly affect virus entry [72,73,74]. |
| A701V | ✔ | . | . | . | S2 Domain | Near Furin Cleavage Site | It boosts viral transmission and aids in viral fitness [87]. |
| N764K | . | . | . | ✔ | S2 Domain | Near Furin Cleavage Site | It provides two cleavage sites for SKI-1/S1P serine protease and increases infectivity [38,77]. |
| D796Y | . | . | . | ✔ | S2 Domain | Near Furin Cleavage Site | It is reported to have a potential impact on protease binding and stabilization of the spike trimer [78,79]. |
| N856K | . | . | . | ✔ | S2 Domain | Near Furin Cleavage Site | It impairs infectivity and provides potential sites for cleavage for serine SKI-1/S1P protease [38,77]. |
| D950N | . | . | ✔ | . | S2 Domain | HR1 | Its particular placement in the trimer interface suggests that it may play a part in changing the behavior of the spike protein [80]. |
| Q954H | . | . | . | ✔ | S2 Domain | HR1 | It improves fusion activity and has a possible contribution toward higher infectivity [81]. |
| N969K | . | . | . | ✔ | S2 Domain | HR1 | It may cause a possible displacement in HR2′s backbone and is reported to impair activity [18,38]. |
| L981F | . | . | . | ✔ | S2 Domain | HR1 | It affects the virus’ binding to the ACE2 receptor and enhances viral spike protein induced infections and syncytia formation [38,79]. |
| T1027I | . | ✔ | . | . | S2 Domain | Trimerization interface | It may be a significant contributor toward enhanced virulence and diminished susceptibility to serum and monoclonal antibodies [32]. |
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, S.; Fogagnolo, A.; Campo, G.; Zucchetti, O.; Verri, M.; Ottaviani, I.; Tunstall, T.; Grasso, S.; Scaramuzzo, V.; Murgolo, F.; et al. Markers of endothelial and epithelial pulmonary injury in mechanically ventilated COVID-19 ICU patients. Crit. Care 2021, 25, 74. [Google Scholar] [CrossRef] [PubMed]
- Contoli, M.; Papi, A.; Tomassetti, L.; Rizzo, P.; Sega, F.V.D.; Fortini, F.; Torsani, F.; Morandi, L.; Ronzoni, L.; Zucchetti, O.; et al. Blood interferon-α levels and severity, outcomes, and inflammatory profiles in hospitalized COVID-19 patients. Front. Immunol. 2021, 12, 648004. [Google Scholar] [CrossRef] [PubMed]
- Sega, F.V.D.; Fortini, F.; Spadaro, S.; Ronzoni, L.; Zucchetti, O.; Manfrini, M.; Mikus, E.; Fogagnolo, A.; Torsani, F.; Pavasini, R.; et al. Time course of endothelial dysfunction markers and mortality in COVID-19 patients: A pilot study. Clin. Transl. Med. 2021, 11, e283. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Mejdani, M.; Haddadi, K.; Pham, C.; Mahadevan, R. SARS-CoV-2 receptor-binding mutations and antibody contact sites. Antib. Ther. 2021, 4, 149–158. [Google Scholar] [CrossRef]
- Weissman, D.; Alameh, M.-G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 2021, 29, 23–31. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Guruprasad, K. Mutations in human SARS-CoV-2 spike proteins, potential drug binding and epitope sites for COVID-19 therapeutics development. Curr. Res. Struct. Biol. 2022, 4, 41–50. [Google Scholar] [CrossRef]
- COVID-19 Map—Johns Hopkins Coronavirus Resource Center. 2022. Available online: https://coronavirus.jhu.edu/map.html (accessed on 4 August 2022).
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. 2019. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 4 August 2022).
- Peters, M.H.; Bastidas, O.; Kokron, D.S.; Henze, C.E. Static all-atom energetic mappings of the SARS-Cov-2 spike protein and dynamic stability analysis of “Up” versus “Down” protomer states. PLoS ONE 2020, 15, e0241168. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Yahi, N.; Chahinian, H.; Fantini, J. Infection-enhancing anti-SARS-CoV-2 antibodies recognize both the original Wuhan/D614G strain and Delta variants. A potential risk for mass vaccination? J. Infect. 2021, 83, 607–635. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S.; Arrigo, N.; Bonet, J.; Xu, Z. Mutational hotspot in the SARS-CoV-2 Spike protein N-terminal domain conferring immune escape potential. Viruses 2021, 13, 2114. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, K.; Singh, P.; Bhargava, A.; Negi, S.S.; Sharma, P.; Bhise, M.; Tripathi, M.K.; Jindal, A.; Nagarkar, N.M. Genomic characterization unravelling the causative role of SARS-CoV-2 Delta variant of lineage B. 1.617. 2 in 2nd wave of COVID-19 pandemic in Chhattisgarh, India. Microb. Pathog. 2022, 164, 105404. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e16. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H.; et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 2021, 29, 44–57.e9. [Google Scholar] [CrossRef]
- Choi, H.; Chatterjee, P.; Hwang, M.; Lichtfouse, E.; Sharma, V.K.; Jinadatha, C. The viral phoenix: Enhanced infectivity and immunity evasion of SARS-CoV-2 variants. Environ. Chem. Lett. 2021, 20, 1539–1544. [Google Scholar] [CrossRef]
- Janik, E.; Niemcewicz, M.; Podogrocki, M.; Majsterek, I.; Bijak, M. The emerging concern and interest SARS-CoV-2 variants. Pathogens 2021, 10, 633. [Google Scholar] [CrossRef]
- Ginex, T.; Marco-Marín, C.; Wieczór, M.; Mata, C.P.; Krieger, J.; Ruiz-Rodriguez, P.; López-Redondo, M.L.; Francés-Gómez, C.; Melero, R.; Sánchez-Sorzano, C.; et al. The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation. PLoS Pathog. 2022, 18, e1010631. [Google Scholar] [CrossRef]
- Alkhatib, M.; Salpini, R.; Carioti, L.; Ambrosio, F.A.; D’Anna, S.; Duca, L.; Costa, G.; Bellocchi, M.C.; Piermatteo, L.; Artese, A.; et al. Update on SARS-CoV-2 omicron variant of concern and its peculiar mutational profile. Microbiol. Spectr. 2022, 10, e02732-21. [Google Scholar] [CrossRef]
- Cathcart, A.L.; Havenar-Daughton, C.; Lempp, F.A.; Ma, D.; Schmid, M.A.; Agostini, M.L.; Guarino, B.; Di iulio, J.; Rosen, L.E.; Tucker, H.; et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. BioRxiv 2022. in pre-print. [Google Scholar] [CrossRef]
- Haslwanter, D.; Dieterle, M.E.; Wec, A.Z.; O’Brien, C.M.; Sakharkar, M.; Florez, C.; Tong, K.; Rappazzo, C.G.; Lasso, G.; Vergnolle, O.; et al. A combination of receptor-binding domain and N-terminal domain neutralizing antibodies limits the generation of SARS-CoV-2 spike neutralization-escape mutants. Mbio 2021, 12, e02473-21. [Google Scholar] [CrossRef] [PubMed]
- Javanmardi, K.; Segall-Shapiro, T.H.; Chou, C.W.; Boutz, D.R.; Olsen, R.J.; Xie, X.; Xia, H.; Shi, P.Y.; Johnson, C.D.; Annapareddy, A.; et al. Antibody escape and cryptic cross-domain stabilization in the SARS-CoV-2 Omicron spike protein. BioRxiv 2022. in pre-print. [Google Scholar] [CrossRef] [PubMed]
- Redd, A.D.; Nardin, A.; Kared, H.; Bloch, E.M.; Pekosz, A.; Laeyendecker, O.; Abel, B.; Fehlings, M.; Quinn, T.C.; Tobian, A.A. CD8+ T-cell responses in COVID-19 convalescent individuals target conserved epitopes from multiple prominent SARS-CoV-2 circulating variants. Open Forum Infect. Dis. 2021, 8, ofab143. [Google Scholar] [CrossRef]
- Das, N.C.; Chakraborty, P.; Bayry, J.; Mukherjee, S. In silico analyses on the comparative potential of therapeutic human monoclonal antibodies against newly emerged SARS-CoV-2 variants bearing mutant spike protein. Front. Immunol. 2021, 12, 782506. [Google Scholar] [CrossRef]
- Shen, L.; Triche, T.J.; Bard, J.D.; Biegel, J.A.; Judkins, A.R.; Gai, X. Spike Protein NTD mutation G142D in SARS-CoV-2 Delta VOC lineages is associated with frequent back mutations, increased viral loads, and immune evasion. medRxiv 2021. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Pathirana, P.N.; Nguyen, T.; Nguyen, Q.V.H.; Bhatti, A.; Nguyen, D.C.; Nguyen, D.T.; Nguyen, N.D.; Creighton, D.; Abdelrazek, M. Genomic mutations and changes in protein secondary structure and solvent accessibility of SARS-CoV-2 (COVID-19 virus). Sci. Rep. 2021, 11, 3487. [Google Scholar] [CrossRef]
- Süt, B.B. Structural analysis of novel amino acid substitutions in SARS-CoV-2 spike protein receptor-binding domain. Hacet. J. Biol. Chem. 2021, 49, 367–374. [Google Scholar] [CrossRef]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.A.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B. 1.1. 7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A.; Simons, L.M.; Adewumi, O.M.; Fowotade, A.A.; Omoruyi, E.C.; Adeniji, J.A.; Dean, T.J.; Zayas, J.; Bhimalli, P.P.; Ash, M.K.; et al. Coincident rapid expansion of two SARS-CoV-2 lineages with enhanced infectivity in Nigeria. medRxiv 2021. [Google Scholar] [CrossRef]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- McEwen, A.E.; Cohen, S.; Bryson-Cahn, C.; Liu, C.; Pergam, S.A.; Lynch, J.; Schippers, A.; Strand, K.; Whimbey, E.; Mani, N.S.; et al. Variants of concern are overrepresented among postvaccination breakthrough infections of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Washington State. Clin. Infect. Dis. 2022, 74, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.; Zhu, Q.; Wang, K.; Chen, R.; Feng, R.; Jia, Z.; et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e13. [Google Scholar] [CrossRef]
- Pastorio, C.; Zech, F.; Noettger, S.; Jung, C.; Jacob, T.; Sanderson, T.; Sparrer, K.M.; Kirchhoff, F. Determinants of Spike infectivity, processing, and neutralization in SARS-CoV-2 Omicron subvariants BA. 1 and BA. 2. Cell Host Microbe 2022, 30, 1255–1268. [Google Scholar] [CrossRef]
- Umair, M.; Ikram, A.; Salman, M.; Badar, N.; Haider, S.A.; Rehman, Z.; Ammar, M.; Rana, M.S.; Ali, Q. Detection and whole-genome sequencing of SARS-CoV-2 B. 1.617. 2 and B. 1.351 variants of concern from Pakistan during the COVID-19 third wave. medRxiv 2021. [Google Scholar] [CrossRef]
- Barton, M.I.; MacGowan, S.A.; Kutuzov, M.A.; Dushek, O.; Barton, G.J.; van der Merwe, P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife 2021, 10, e70658. [Google Scholar] [CrossRef]
- Fratev, F. N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with Both hACE2 and human-derived antibody: A free energy of perturbation retrospective study. J. Chem. Inf. Model. 2021, 61, 6079–6084. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Q.; Ge, J.; Ren, W.; Zhang, R.; Lan, J.; Ju, B.; Su, B.; Yu, F.; Chen, P.; et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity 2021, 54, 1611–1621.e5. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Wang, P.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.; et al. Increased resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7 to antibody neutralization. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Bernard, D.W.; Davis, J.J.; Shukla, M.; Nguyen, M.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; et al. Molecular architecture of early dissemination and massive second wave of the SARS-CoV-2 virus in a major metropolitan area. MBio 2020, 11, e02707-20. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Deng, Y.-Q.; Ye, Q.; Cao, L.; Sun, C.-Y.; Fan, C.; Huang, W.; Sun, S.; Sun, Y.; Zhu, L.; et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020, 369, 1505–1509. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.-W.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602, 676–681. [Google Scholar] [CrossRef]
- Martinez, D.R.; Schäfer, A.; Gobeil, S.; Li, D.; De la Cruz, G.; Parks, R.; Lu, X.; Barr, M.; Stalls, V.; Janowska, K.; et al. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. 2021, 14, eabj7125. [Google Scholar] [CrossRef]
- Das, C.; Hazarika, P.J.; Deb, A.; Joshi, P.; Das, D.; Mattaparthi, V.S.K. Effect of double mutation (L452R and E484Q) in RBD of SPIKE Protein on its interaction with ACE2 receptor protein. Biointerface Res. Appl. Chem. 2022, 13, 97. [Google Scholar] [CrossRef]
- Augusto, G.; Mohsen, M.O.; Zinkhan, S.; Liu, X.; Vogel, M.; Bachmann, M.F. In vitro data suggest that Indian delta variant B. 1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy 2022, 77, 111–117. [Google Scholar] [CrossRef]
- Rostami, N.; Choupani, E.; Hernandez, Y.; Arab, S.S.; Jazayeri, S.M.; Gomari, M.M. SARS-CoV-2 spike evolutionary behaviors; simulation of N501Y mutation outcomes in terms of immunogenicity and structural characteristic. J. Cell. Biochem. 2022, 123, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.A.T.M.; Kemp, S.A.; Datir, R.; Saito, A.; Meng, B.; Rakshit, P.; Takaori-Kondo, A.; Kosugi, Y.; Uriu, K.; Kimura, I.; et al. SARS-CoV-2 B. 1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J. Infect. Dis. 2021, 224, 989–994. [Google Scholar] [CrossRef]
- Chakraborty, S. E484K and N501Y SARS-CoV 2 spike mutants increase ACE2 recognition but reduce affinity for neutralizing antibody. Int. Immunopharmacol. 2022, 102, 108424. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-T.; Huang, W.-H.; Liao, T.-L.; Hsiao, T.-H.; Chuang, H.-N.; Liu, P.-Y. SARS-CoV-2 E484K mutation narrative review: Epidemiology, immune escape, clinical implications, and future considerations. Infect. Drug Resist. 2022, 15, 373. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Kasry, A.; Amin, M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med. Drug Discov. 2021, 10, 100086. [Google Scholar] [CrossRef]
- Santos, J.C.; Passos, G.A. The high infectivity of SARS-CoV-2 B. 1.1. 7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. BioRxiv 2021. in pre-print. [Google Scholar] [CrossRef]
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife 2021, 10, e69091. [Google Scholar] [CrossRef]
- Teruel, N.; Mailhot, O.; Najmanovich, R.J. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 Spike protein variants. PLoS Comput. Biol. 2021, 17, e1009286. [Google Scholar] [CrossRef]
- Di Giacomo, S.; Mercatelli, D.; Rakhimov, A.; Giorgi, F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021, 93, 5638–5643. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022, 126, 102779. [Google Scholar] [CrossRef]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361.e6. [Google Scholar] [CrossRef] [PubMed]
- Berger, I.; Schaffitzel, C. The SARS-CoV-2 spike protein: Balancing stability and infectivity. Cell Res. 2020, 30, 1059–1060. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Edwards, R.J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S.M.C.; Kopp, M.; Li, D.; Parks, R.; Hsu, A.L.; et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020, 27, 925–933. [Google Scholar] [CrossRef]
- Al-Zyoud, W.; Haddad, H. Potential linear B-cells epitope change to a helix structure in the spike of Omicron 21L or BA. 2 predicts increased SARS-CoV-2 antibodies evasion. Virology 2022, 573, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Han, W.; Li, J.; Xu, S.; Wang, Y.; Xu, C.; Li, Z.; Wang, Y.; Zhang, C.; Huang, Z.; et al. Molecular basis of receptor binding and antibody neutralization of Omicron. Nature 2022, 604, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Evans, J.P.; Qu, P.; Faraone, J.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Zhou, T.; Lozanski, G.; Mallampalli, R.; et al. Neutralization and stability of SARS-CoV-2 Omicron variant. BioRxiv 2021, preprint. [Google Scholar] [CrossRef]
- Ozono, S.; Zhang, Y.; Ode, H.; Sano, K.; Tan, T.S.; Imai, K.; Miyoshi, K.; Kishigami, S.; Ueno, T.; Iwatani, Y.; et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021, 12, 848. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.; Xiao, T.; Lu, J.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rits-Volloch, S.; Zhu, H.; Woosley, A.N.; et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021, 372, 525–530. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. BioRxiv 2021. [Google Scholar] [CrossRef]
- Lista, M.J.; Winstone, H.; Wilson, H.D.; Dyer, A.; Pickering, S.; Galao, R.P.; De Lorenzo, G.; Cowton, V.M.; Furnon, W.; Suarez, N.; et al. The P681H mutation in the Spike glycoprotein confers type I interferon resistance in the SARS-CoV-2 alpha (B. 1.1. 7) variant. BioRxiv 2021. [Google Scholar] [CrossRef]
- Lubinski, B.; Fernandes, M.H.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B. 1.1. 7 (Alpha) spike. iScience 2022, 25, 103589. [Google Scholar] [CrossRef] [PubMed]
- Boehm, E.; Kronig, I.; Neher, R.A.; Eckerle, I.; Vetter, P.; Kaiser, L. Novel SARS-CoV-2 variants: The pandemics within the pandemic. Clin. Microbiol. Infect. 2021, 27, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; Jaimes, J.A.; Whittaker, G. Intrinsic furin-mediated cleavability of the spike S1/S2 site from SARS-CoV-2 variant B. 1.529 (Omicron). BioRxiv 2022. [Google Scholar] [CrossRef]
- Maaroufi, H. The N764K and N856K mutations in SARS-CoV-2 Omicron BA. 1 S protein generate potential cleavage sites for SKI-1/S1P protease. BioRxiv 2022. [Google Scholar] [CrossRef]
- Ni, D.; Lau, K.; Turelli, P.; Raclot, C.; Beckert, B.; Nazarov, S.; Pojer, F.; Myasnikov, A.; Stahlberg, H.; Trono, D. Structural analysis of the spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion. BioRxiv 2021. [Google Scholar] [CrossRef]
- Ou, J.; Lan, W.; Wu, X.; Zhao, T.; Duan, B.; Yang, P.; Ren, Y.; Quan, L.; Zhao, W.; Seto, D.; et al. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Target. Ther. 2022, 7, 138. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, R.; Lo, M.; Saha, R.; Dutta, S.; Chawla-Sarkar, M. S glycoprotein diversity of the Omicron variant. MedRxiv 2021. [Google Scholar] [CrossRef]
- Yang, K.; Wang, C.; White, K.I.; Pfuetzner, R.A.; Esquivies, L.; Brunger, A.T. Structural conservation among variants of the SARS-CoV-2 spike postfusion bundle. Proc. Natl. Acad. Sci. USA 2022, 119, e2119467119. [Google Scholar] [CrossRef]
- Kemp, S.A.; Meng, B.; Ferriera, I.A.; Datir, R.; Harvey, W.T.; Papa, G.; Lytras, S.; Collier, D.A.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70. BioRxiv 2021, 35, 13. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef] [PubMed]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Harari, D.; Chiaravalli, J.; Meyer, B.; Rudich, Y.; Li, C.; Marton, I.; et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021, 6, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Suppiah, J.; Kamel, K.A.; Mohd-Zawawi, Z.; Afizan, M.A.; Yahya, H.; Md-Hanif, S.A.; Thayan, R. SHORT COMMUNICATION Phylogenomic analysis of SARS-CoV-2 from third wave clusters in Malaysia reveals dominant local lineage B. 1.524 and persistent spike mutation A701V. Trop. Biomed. 2021, 38, 289–293. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; La Scola, B.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022, 94, 2019–2025. [Google Scholar] [CrossRef]
- Hoffmann, M.; Krüger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.-S.; Winkler, M.S.; et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 2022, 185, 447–456.e11. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asif, A.; Ilyas, I.; Abdullah, M.; Sarfraz, S.; Mustafa, M.; Mahmood, A. The Comparison of Mutational Progression in SARS-CoV-2: A Short Updated Overview. J. Mol. Pathol. 2022, 3, 201-218. https://doi.org/10.3390/jmp3040018
Asif A, Ilyas I, Abdullah M, Sarfraz S, Mustafa M, Mahmood A. The Comparison of Mutational Progression in SARS-CoV-2: A Short Updated Overview. Journal of Molecular Pathology. 2022; 3(4):201-218. https://doi.org/10.3390/jmp3040018
Chicago/Turabian StyleAsif, Abeer, Iqra Ilyas, Mohammad Abdullah, Sadaf Sarfraz, Muhammad Mustafa, and Arif Mahmood. 2022. "The Comparison of Mutational Progression in SARS-CoV-2: A Short Updated Overview" Journal of Molecular Pathology 3, no. 4: 201-218. https://doi.org/10.3390/jmp3040018






