Novel Pseudomonas Species Prevent the Growth of the Phytopathogenic Fungus Aspergillus flavus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Growth Conditions
2.2. Phylogenetic Analysis of BJa3 and MCal1 Isolates
2.3. Detection of VOCs Antagonistic Activity In Vitro
2.4. Detection of Soluble Antagonist Compounds
2.5. Statistical Data Analysis
2.6. Morphological Characterization of A. flavus after VOC Exposure of BJa3, MCal1, and DH5α by Confocal Laser Scanning Microscopy (CLSM)
2.7. Bacterial VOCs Identification by Solid-Phase Microextraction Coupled to Gas Chromatography-Mass Spectrometry (SPME/GC–MS)
2.8. Bacterial Volatilome Analysis
3. Results and Discussion
3.1. BJa3 and MCal1 Produce Volatile Compounds with Antifungal Activity
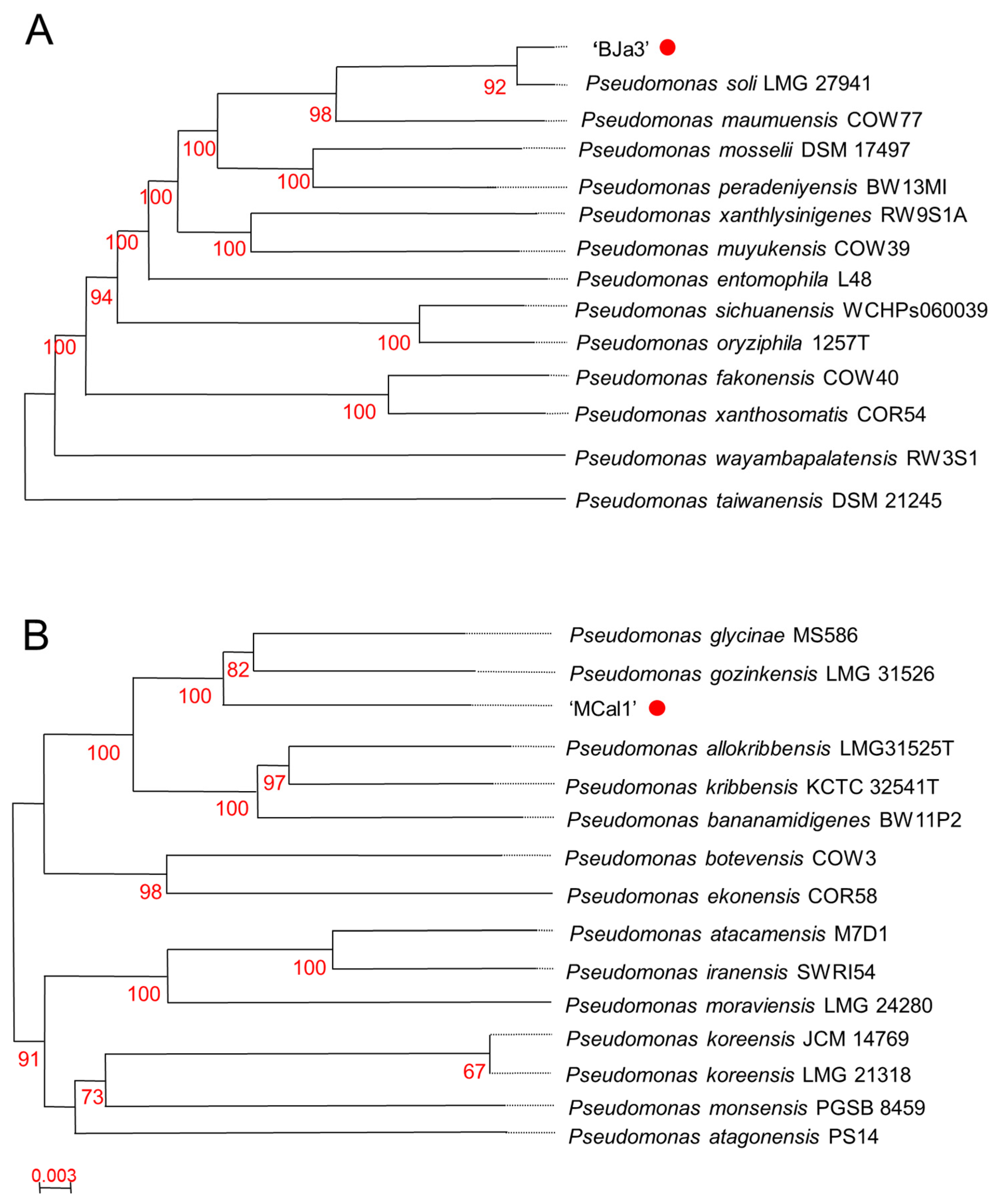
3.2. The Strain MCal1 Represents a Novel Species within the Pseudomonas Genus, while BJa3 Is Categorized within the Pseudomonas Soli Species
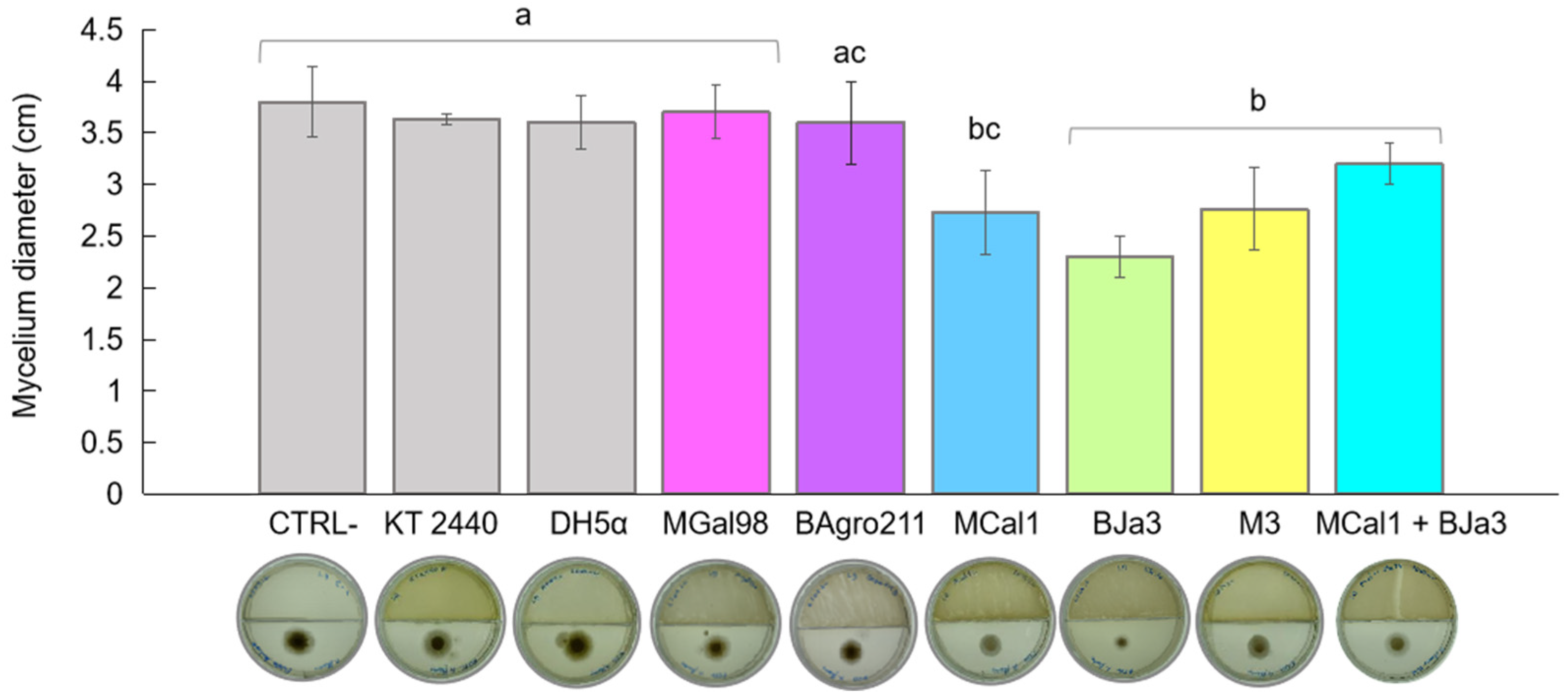
3.3. MCal1 and BJa3 Also Exhibit Antimicrobial Effects on A. flavus through Diffusible Compounds Production
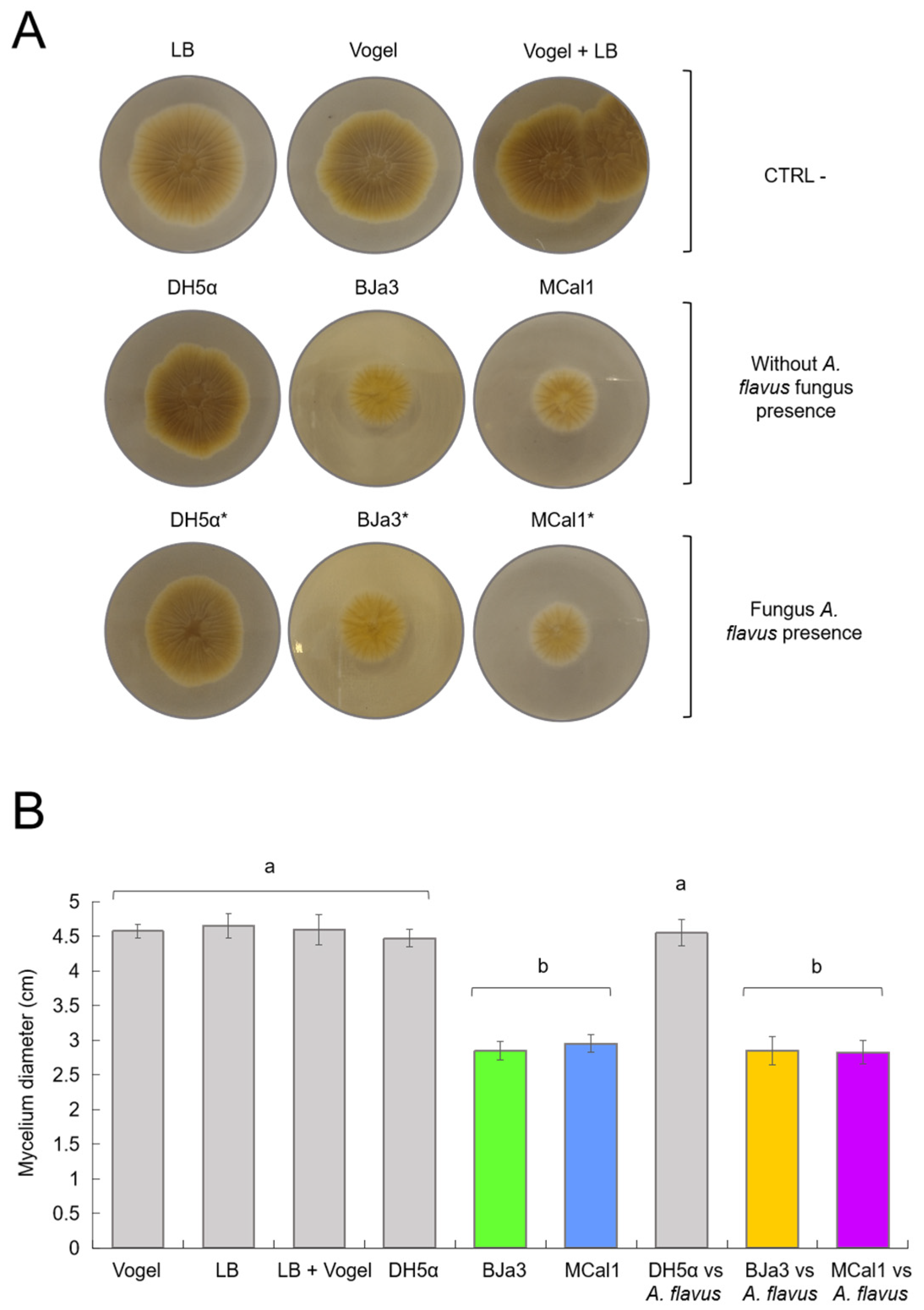
3.4. VOCs Produced by BJa3 and MCal1 Strains Affect the Morphology of A. flavus
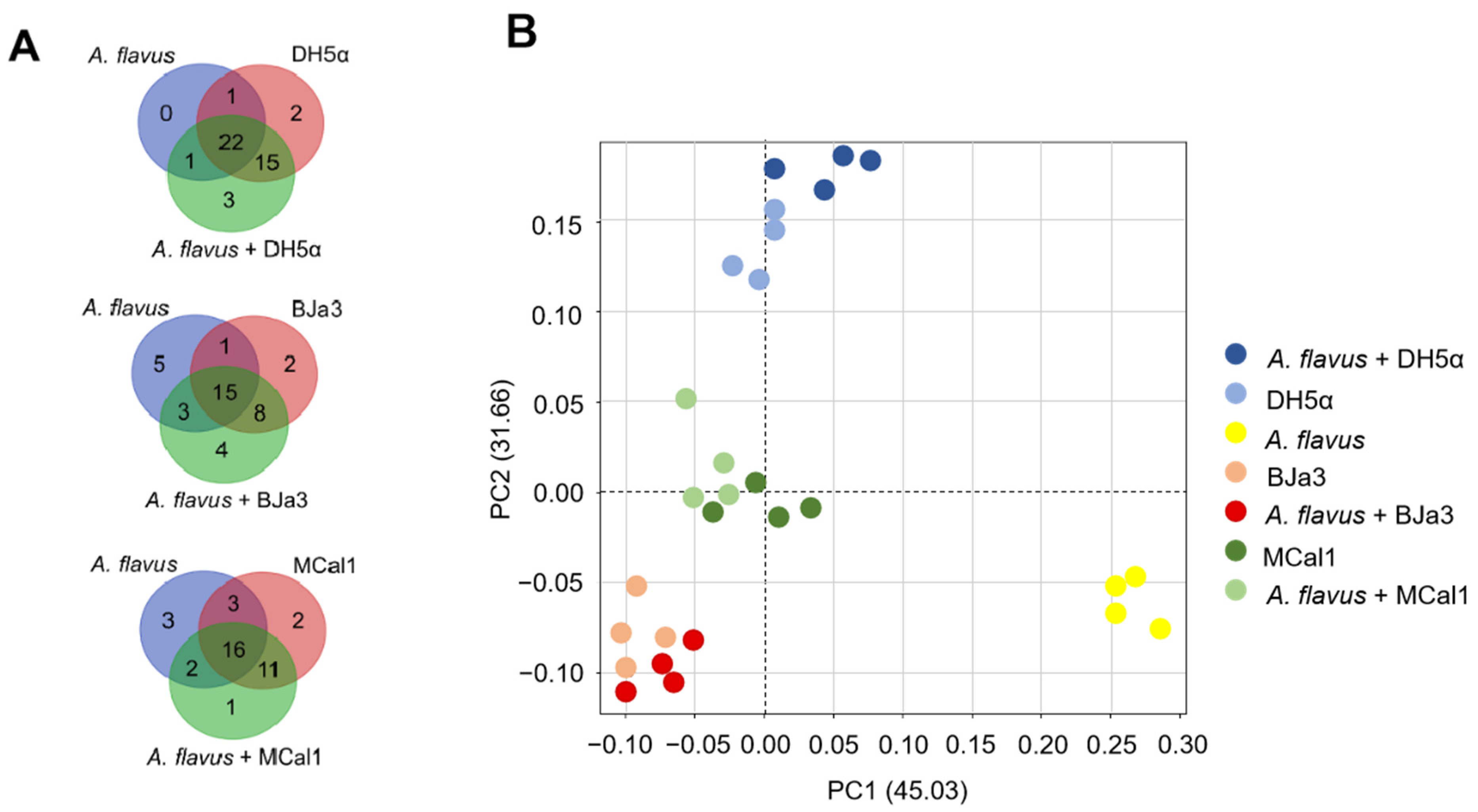
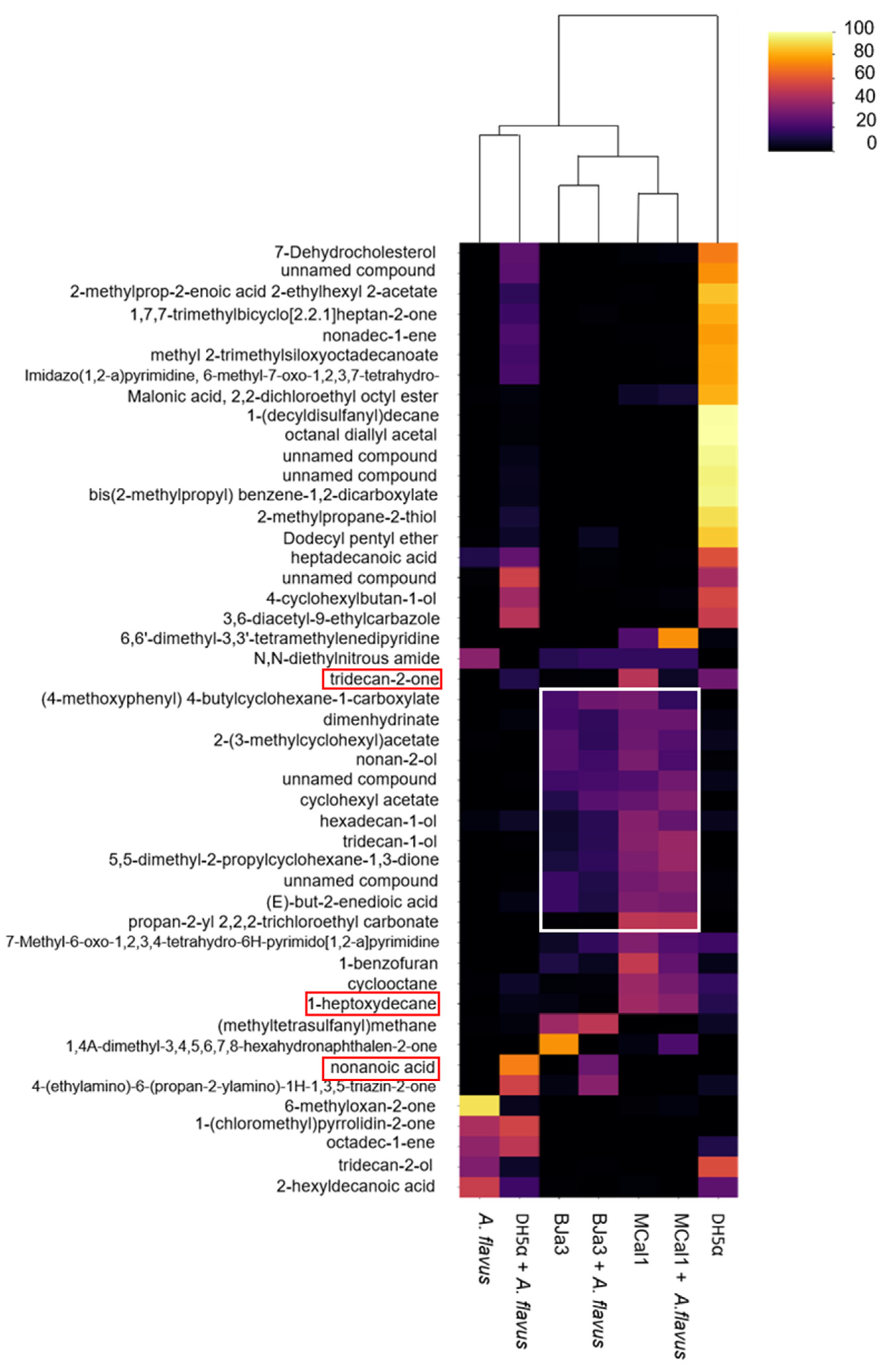
3.5. Strains BJa3, MCal1, and DH5α Produce VOCs That Are Species-Specific and Pseudomonas Strains Produce Compounds with Antifungal Potential
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdallah, M.F.; Audenaert, K.; Saeger, S.D.; Houbraken, J. Revisiting an Aspergillus Flavus Strain Isolated from an Egyptian Sugarcane Field in 1930. Microorganisms 2020, 8, 1633. [Google Scholar] [CrossRef]
- Coomes, T.J.; Sanders, J.C. The Detection and Estimation of Aflatoxin in Groundnuts and Groundnut Materials. Part I. Paper-Chromatographic Procedure. Analyst 1963, 88, 209. [Google Scholar] [CrossRef]
- Hartley, R.D.; Nesbitt, B.F.; O’Kelly, J. Toxic Metabolites of Aspergillus Flavus. Nature 1963, 198, 1056–1058. [Google Scholar] [CrossRef]
- Viaro, H.P.; da Silva, J.J.; de Souza Ferranti, L.; Bordini, J.G.; Massi, F.P.; Fungaro, M.H.P. The First Report of A. Novoparasiticus, A. Arachidicola and A. Pseudocaelatus in Brazilian Corn Kernels. Int. J. Food Microbiol. 2017, 243, 46–51. [Google Scholar] [CrossRef]
- Silva, J.J.; Puel, O.; Lorber, S.; Ferranti, L.S.; Ortiz, L.F.; Taniwaki, M.H.; Iamanaka, B.T.; Fungaro, M.H.P. Occurrence and Diversity of Aspergillus in Commercial Yerba Mate Elaborated for the Brazilian Beverage ‘Chimarrão’. Food Res. Int. 2019, 121, 940–946. [Google Scholar] [CrossRef]
- Iamanaka, B.T.; de Souza Lopes, A.; Martins, L.M.; Frisvad, J.C.; Medina, A.; Magan, N.; Sartori, D.; Massi, F.P.; Fungaro, M.H.P.; Taniwaki, M.H. Aspergillus Section Flavi Diversity and the Role of A. novoparasiticus in Aflatoxin Contamination in the Sugarcane Production Chain. Int. J. Food Microbiol. 2019, 293, 17–23. [Google Scholar] [CrossRef]
- Ismaiel, A.; Papenbrock, J. Mycotoxins: Producing Fungi and Mechanisms of Phytotoxicity. Agriculture 2015, 5, 492–537. [Google Scholar] [CrossRef]
- Mitchell, N.J.; Bowers, E.; Hurburgh, C.; Wu, F. Potential Economic Losses to the US Corn Industry from Aflatoxin Contamination. Food Addit. Contam. Part A 2016, 33, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Umesha, S.; Manukumar, H.M.G.; Chandrasekhar, B.; Shivakumara, P.; Shiva Kumar, J.; Raghava, S.; Avinash, P.; Shirin, M.; Bharathi, T.R.; Rajini, S.B.; et al. A Flatoxins and Food Pathogens: Impact of Biologically Active Aflatoxins and Their Control Strategies: Aflatoxins and Food Pathogens. J. Sci. Food Agric. 2017, 97, 1698–1707. [Google Scholar] [CrossRef]
- Lemfack, M.-C.; Bahl, H.; Piechulla, B.; Magnus, N. The Domain of Bacteria and Their Volatile Metabolic Potential. In Bacterial Volatile Compounds as Mediators of Airborne Interactions; Ryu, C.-M., Weisskopf, L., Piechulla, B., Eds.; Springer: Singapore, 2020; pp. 1–38. ISBN 9789811572920. [Google Scholar]
- Tilocca, B.; Cao, A.; Migheli, Q. Scent of a Killer: Microbial Volatilome and Its Role in the Biological Control of Plant Pathogens. Front. Microbiol. 2020, 11, 41. [Google Scholar] [CrossRef]
- Mari, M.; Bautista-Baños, S.; Sivakumar, D. Decay Control in the Postharvest System: Role of Microbial and Plant Volatile Organic Compounds. Postharvest Biol. Technol. 2016, 122, 70–81. [Google Scholar] [CrossRef]
- Qin, X.; Xiao, H.; Cheng, X.; Zhou, H.; Si, L. Hanseniaspora Uvarum Prolongs Shelf Life of Strawberry via Volatile Production. Food Microbiol. 2017, 63, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Toffano, L.; Fialho, M.B.; Pascholati, S.F. Potential of Fumigation of Orange Fruits with Volatile Organic Compounds Produced by Saccharomyces Cerevisiae to Control Citrus Black Spot Disease at Postharvest. Biol. Control 2017, 108, 77–82. [Google Scholar] [CrossRef]
- Peñuelas, J.; Asensio, D.; Tholl, D.; Wenke, K.; Rosenkranz, M.; Piechulla, B.; Schnitzler, J.P. Biogenic Volatile Emissions from the Soil: Biogenic Volatile Emissions from the Soil. Plant Cell Environ. 2014, 37, 1866–1891. [Google Scholar] [CrossRef]
- Schenkel, D.; Deveau, A.; Niimi, J.; Mariotte, P.; Vitra, A.; Meisser, M.; Buttler, A.; Splivallo, R. Linking Soil’s Volatilome to Microbes and Plant Roots Highlights the Importance of Microbes as Emitters of Belowground Volatile Signals. Environ. Microbiol. 2019, 21, 3313–3327. [Google Scholar] [CrossRef]
- Hernández-León, R.; Rojas-Solís, D.; Contreras-Pérez, M.; del Carmen Orozco-Mosqueda, M.; Macías-Rodríguez, L.I.; Reyes-de la Cruz, H.; Valencia-Cantero, E.; Santoyo, G. Characterization of the Antifungal and Plant Growth-Promoting Effects of Diffusible and Volatile Organic Compounds Produced by Pseudomonas Fluorescens Strains. Biol. Control 2015, 81, 83–92. [Google Scholar] [CrossRef]
- Guo, J.; Xu, Y.; Liang, S.; Zhou, Z.; Zhang, C.; Li, K.; Peng, X.; Qin, S.; Xing, K. Antifungal Activity of Volatile Compounds from Bacillus tequilensis XK29 against Botrytis Cinerea Causing Gray Mold on Cherry Tomatoes. Postharvest Biol. Technol. 2023, 198, 112239. [Google Scholar] [CrossRef]
- Wallace, R.L.; Hirkala, D.L.; Nelson, L.M. Mechanisms of Action of Three Isolates of Pseudomonas Fluorescens Active against Postharvest Grey Mold Decay of Apple during Commercial Storage. Biological Control 2018, 117, 13–20. [Google Scholar] [CrossRef]
- Raza, W.; Ling, N.; Liu, D.; Wei, Z.; Huang, Q.; Shen, Q. Volatile Organic Compounds Produced by Pseudomonas Fluorescens WR-1 Restrict the Growth and Virulence Traits of Ralstonia Solanacearum. Microbiol. Res. 2016, 192, 103–113. [Google Scholar] [CrossRef]
- Gandía, M.; Kakar, A.; Giner-Llorca, M.; Holzknecht, J.; Martínez-Culebras, P.; Galgóczy, L.; Marx, F.; Marcos, J.F.; Manzanares, P. Potential of Antifungal Proteins (AFPs) to Control Penicillium Postharvest Fruit Decay. JoF 2021, 7, 449. [Google Scholar] [CrossRef]
- Ling, L.; Jiang, K.; Cheng, W.; Wang, Y.; Pang, M.; Luo, H.; Lu, L.; Gao, K.; Tu, Y. Biocontrol of Volatile Organic Compounds Obtained from Bacillus subtilis CL2 against Aspergillus Flavus in Peanuts during Storage. Biol. Control 2022, 176, 105094. [Google Scholar] [CrossRef]
- Natarajan, S.; Balachandar, D.; Senthil, N.; Velazhahan, R.; Paranidharan, V. Volatiles of Antagonistic Soil Yeasts Inhibit Growth and Aflatoxin Production of Aspergillus Flavus. Microbiol. Res. 2022, 263, 127150. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Ishihara, A.; Nakajima, H. Isolation of Anteiso-C 17, Iso-C 17, Iso-C 16, and Iso-C 15 Bacillomycin D from Bacillus amyloliquefaciens SD-32 and Their Antifungal Activities against Plant Pathogens. J. Agric. Food Chem. 2014, 62, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Li, B.; Zhang, N.; Waseem, R.; Shen, Q.; Huang, Q. Production of Bacillomycin- and Macrolactin-Type Antibiotics by Bacillus amyloliquefaciens NJN-6 for Suppressing Soilborne Plant Pathogens. J. Agric. Food Chem. 2012, 60, 2976–2981. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Li, N.; Li, B.; Wang, S.; Wang, Y.; Meng, X.; Li, B.; Cao, K.; Hu, T. An Endophytic Strain Bacillus velezensis JZ51 Controlled Pink Mold Rot of Postharvest Apple Fruit via Antagonistic Action and Disease Resistance Induction. Postharvest Biol. Technol. 2024, 210, 112793. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile Weapons for Plant Disease Biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Vollenbroich, D.; Özel, M.; Vater, J.; Kamp, R.M.; Pauli, G. Mechanism of Inactivation of Enveloped Viruses by the Biosurfactant Surfactin from Bacillus subtilis. Biologicals 1997, 25, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Piro, V.C.; Faoro, H.; Weiss, V.A.; Steffens, M.B.; Pedrosa, F.O.; Souza, E.M.; Raittz, R.T. FGAP: An Automated Gap Closing Tool. BMC Res. Notes 2014, 7, 371. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Y.; Zhao, C.-A.; Liu, C.-J.; Xu, X.-F. Endophytic Fungi Diversity of Aquatic/Riparian Plants and Their Antifungal Activity in Vitro. J. Microbiol. 2010, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Ugolini, L.; Lazzeri, L.; Mari, M. Production of Volatile Organic Compounds by Aureobasidium Pullulans as a Potential Mechanism of Action against Postharvest Fruit Pathogens. Biol. Control 2015, 81, 8–14. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Nachar, N. The Mann-Whitney U: A Test for Assessing Whether Two Independent Samples Come from the Same Distribution. Tutor. Quant. Methods Psychol. 2008, 4, 13–20. [Google Scholar] [CrossRef]
- Chiniquy, D.; Barnes, E.M.; Zhou, J.; Hartman, K.; Li, X.; Sheflin, A.; Pella, A.; Marsh, E.; Prenni, J.; Deutschbauer, A.M.; et al. Microbial Community Field Surveys Reveal Abundant Pseudomonas Population in Sorghum Rhizosphere Composed of Many Closely Related Phylotypes. Front. Microbiol. 2021, 12, 598180. [Google Scholar] [CrossRef]
- Castaldi, S.; Masi, M.; Sautua, F.; Cimmino, A.; Isticato, R.; Carmona, M.; Tuzi, A.; Evidente, A. Pseudomonas Fluorescens Showing Antifungal Activity against Macrophomina Phaseolina, a Severe Pathogenic Fungus of Soybean, Produces Phenazine as the Main Active Metabolite. Biomolecules 2021, 11, 1728. [Google Scholar] [CrossRef]
- Castro Tapia, M.P.; Madariaga Burrows, R.P.; Ruiz Sepúlveda, B.; Vargas Concha, M.; Vera Palma, C.; Moya-Elizondo, E.A. Antagonistic Activity of Chilean Strains of Pseudomonas Protegens Against Fungi Causing Crown and Root Rot of Wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Gao, J.; Zhang, M.; Xue, J.; Zhang, X. Pseudomonas Aeruginosa Ld-08 Isolated from Lilium Davidii Exhibits Antifungal and Growth-Promoting Properties. PLoS ONE 2022, 17, e0269640. [Google Scholar] [CrossRef] [PubMed]
- Melo, I.; Souza, W.; Silva, L.; Santos, S.; Assalin, M.; Zucchi, T.; Queiroz, S. Antifungal Activity of Pseudomonas Frederiksbergensis CMAA 1323 Isolated from the Antarctic Hair Grass Deschampsia Antarctica. Br. Microbiol. Res. J. 2016, 14, 1–11. [Google Scholar] [CrossRef]
- Müller, T.; Ruppel, S.; Behrendt, U.; Lentzsch, P.; Müller, M.E.H. Antagonistic Potential of Fluorescent Pseudomonads Colonizing Wheat Heads Against Mycotoxin Producing Alternaria and Fusaria. Front. Microbiol. 2018, 9, 2124. [Google Scholar] [CrossRef]
- Kai, M.; Haustein, M.; Molina, F.; Petri, A.; Scholz, B.; Piechulla, B. Bacterial Volatiles and Their Action Potential. Appl. Microbiol. Biotechnol. 2009, 81, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Tyc, O.; Zweers, H.; de Boer, W.; Garbeva, P. Volatiles in Inter-Specific Bacterial Interactions. Front. Microbiol. 2015, 6, 166507. [Google Scholar] [CrossRef]
- Abis, L.; Loubet, B.; Ciuraru, R.; Lafouge, F.; Houot, S.; Nowak, V.; Tripied, J.; Dequiedt, S.; Maron, P.A.; Sadet-Bourgeteau, S. Reduced Microbial Diversity Induces Larger Volatile Organic Compound Emissions from Soils. Sci. Rep. 2020, 10, 6104. [Google Scholar] [CrossRef] [PubMed]
- van Agtmaal, M.; van Os, G.J.; Hol, W.H.G.; Hundscheid, M.P.J.; Runia, W.T.; Hordijk, C.A.; de Boer, W. Legacy Effects of Anaerobic Soil Disinfestation on Soil Bacterial Community Composition and Production of Pathogen-Suppressing Volatiles. Front. Microbiol. 2015, 6, 701. [Google Scholar] [CrossRef]
- Song, G.; Ryu, C.-M. Two Volatile Organic Compounds Trigger Plant Self-Defense against a Bacterial Pathogen and a Sucking Insect in Cucumber under Open Field Conditions. Int. J. Mol. Sci. 2013, 14, 9803–9819. [Google Scholar] [CrossRef]
- Goelen, T.; Vuts, J.; Sobhy, I.S.; Wäckers, F.; Caulfield, J.C.; Birkett, M.A.; Rediers, H.; Jacquemyn, H.; Lievens, B. Identification and Application of Bacterial Volatiles to Attract a Generalist Aphid Parasitoid: From Laboratory to Greenhouse Assays. Pest Manag. Sci. 2021, 77, 930–938. [Google Scholar] [CrossRef]
- Chen, W.-H.; You, F. Smart Greenhouse Control under Harsh Climate Conditions Based on Data-Driven Robust Model Predictive Control with Principal Component Analysis and Kernel Density Estimation. J. Process Control 2021, 107, 103–113. [Google Scholar] [CrossRef]
- Gan, Z.; Zhou, Q.; Zheng, C.; Wang, J. Challenges and Applications of Volatile Organic Compounds Monitoring Technology in Plant Disease Diagnosis. Biosens. Bioelectron. 2023, 237, 115540. [Google Scholar] [CrossRef]
- Glare, T.; Caradus, J.; Gelernter, W.; Jackson, T.; Keyhani, N.; Köhl, J.; Marrone, P.; Morin, L.; Stewart, A. Have Biopesticides Come of Age? Trends Biotechnol. 2012, 30, 250–258. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Fuentes, J.D.; Roulston, T.; Kathilankal, J.C.; Lerdau, M. Effects of Air Pollution on Biogenic Volatiles and Ecological Interactions. Oecologia 2009, 160, 411–420. [Google Scholar] [CrossRef]
- Blande, J.D.; Holopainen, J.K.; Niinemets, Ü. Plant Volatiles in Polluted Atmospheres: Stress Responses and Signal Degradation. Plant Cell Environ. 2014, 37, 1892–1904. [Google Scholar] [CrossRef]
- Insam, H.; Seewald, M.S.A. Volatile Organic Compounds (VOCs) in Soils. Biol. Fertil. Soils 2010, 46, 199–213. [Google Scholar] [CrossRef]
- Rani, A.; Rana, A.; Dhaka, R.K.; Singh, A.P.; Chahar, M.; Singh, S.; Nain, L.; Singh, K.P.; Minz, D. Bacterial Volatile Organic Compounds as Biopesticides, Growth Promoters and Plant-Defense Elicitors: Current Understanding and Future Scope. Biotechnol. Adv. 2023, 63, 108078. [Google Scholar] [CrossRef]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas Putida Group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Saati-Santamaría, Z.; Peral-Aranega, E.; Velázquez, E.; Rivas, R.; García-Fraile, P. Phylogenomic Analyses of the Genus Pseudomonas Lead to the Rearrangement of Several Species and the Definition of New Genera. Biology 2021, 10, 782. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Gomila, M.; Mulet, M.; Zaruma, A.; García-Valdés, E. Past, Present and Future of the Boundaries of the Pseudomonas Genus: Proposal of Stutzerimonas Gen. Nov. Syst. Appl. Microbiol. 2022, 45, 126289. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Klenk, H.-P.; Göker, M. Taxonomic Use of DNA G+C Content and DNA–DNA Hybridization in the Genomic Age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Corallo, B.; Amarelle, V.; Stewart, S.; Pan, D.; Tiscornia, S.; Fabiano, E. Paenibacillus sp. Strain UY79, Isolated from a Root Nodule of Arachis villosa, Displays a Broad Spectrum of Antifungal Activity. Appl. Environ. Microbiol. 2022, 88, e01645-21. [Google Scholar] [CrossRef] [PubMed]
- Remali, J.; Sarmin, N.; Izzah, M.; Ng, C.L.; Tiong, J.J.L.; Aizat, W.M.; Keong, L.K.; Zin, N.M. Genomic Characterization of a New Endophytic Streptomyces Kebangsaanensis Identifies Biosynthetic Pathway Gene Clusters for Novel Phenazine Antibiotic Production. PeerJ 2017, 5, e3738. [Google Scholar] [CrossRef] [PubMed]
- Siupka, P.; Piński, A.; Babicka, D.; Piotrowska-Seget, Z. Genome Mining Revealed a High Biosynthetic Potential for Antifungal streptomyces sp. S-2 Isolated from Black Soot. Int. J. Mol. Sci. 2020, 21, 2558. [Google Scholar] [CrossRef] [PubMed]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.-C.; Dietl, A.-M.; Visca, P.; Haas, H.; et al. Studies of Pseudomonas Aeruginosa Mutants Indicate Pyoverdine as the Central Factor in Inhibition of Aspergillus Fumigatus Biofilm. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.; Sharma, D.; Kumar, P.; Chandra Dubey, R. Biocontrol Potential of Bacillus spp. for Resilient and Sustainable Agricultural Systems. Physiol. Mol. Plant Pathol. 2023, 128, 102173. [Google Scholar] [CrossRef]
- Jenul, C.; Sieber, S.; Daeppen, C.; Mathew, A.; Lardi, M.; Pessi, G.; Hoepfner, D.; Neuburger, M.; Linden, A.; Gademann, K.; et al. Biosynthesis of Fragin Is Controlled by a Novel Quorum Sensing Signal. Nat. Commun. 2018, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Kruijt, M.; Tran, H.; Raaijmakers, J.M. Functional, Genetic and Chemical Characterization of Biosurfactants Produced by Plant Growth-Promoting Pseudomonas Putida 267. J. Appl. Microbiol. 2009, 107, 546–556. [Google Scholar] [CrossRef]
- Nishanth Kumar, S.; Aravind, S.R.; Jacob, J.; Gopinath, G.; Lankalapalli, R.S.; Sreelekha, T.T.; Dileep Kumar, B.S. Pseudopyronine B: A Potent Antimicrobial and Anticancer Molecule Isolated from a Pseudomonas Mosselii. Front. Microbiol. 2016, 7, 1307. [Google Scholar] [CrossRef]
- Martin, H.C.; Ibáñez, R.; Nothias, L.-F.; Boya, P.C.A.; Reinert, L.K.; Rollins-Smith, L.A.; Dorrestein, P.C.; Gutiérrez, M. Viscosin-like Lipopeptides from Frog Skin Bacteria Inhibit Aspergillus Fumigatus and Batrachochytrium Dendrobatidis Detected by Imaging Mass Spectrometry and Molecular Networking. Sci. Rep. 2019, 9, 3019. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Wang, H.; Tan, Z.; Xuan, Z.; Dahar, G.Y.; Li, Q.X.; Miao, W.; Liu, W. Antifungal Mechanism of Bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum Gloeosporioides Penz. Pestic. Biochem. Physiol. 2020, 163, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Vanittanakom, N.; Loeffler, W.; Koch, U.; Jung, G. Fengycin—A Novel Antifungal Lipopeptide Antibiotic Produced by Bacillus subtilis F-29-3. J. Antibiot. 1986, 39, 888–901. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.; Soares, C.; Kozakiewicz, Z.; Paterson, R.R.M.; Lima, N.; Venâncio, A. Identification and Characterization of Aspergillus Flavus and Aflatoxins. Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 527–534. [Google Scholar]
- Boukaew, S.; Plubrukam, A.; Prasertsan, P. Effect of Volatile Substances from Streptomyces Philanthi RM-1-138 on Growth of Rhizoctonia Solani on Rice Leaf. BioControl 2013, 58, 471–482. [Google Scholar] [CrossRef]
- Moore-Landecker, E.; Stotzky, G. Morphological Abnormalities of Fungi Induced by Volatile Microbial Metabolites. Mycologia 1973, 65, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Nagayama, K.; Fujita, K.; Takashima, Y.; Ardin, A.C.; Ooshima, T.; Matsumoto-Nakano, M. Role of ABC Transporter Proteins in Stress Responses of Streptococcus Mutans. Oral Health Dent. Manag. 2014, 13, 359–365. [Google Scholar] [PubMed]
- Rolim, J.M.; Walker, C.; Mezzomo, R.; Muniz, M.F. Antagonism and Effect of Volatile Metabolites of Trichoderma spp. on Cladosporium spp. Floresta Ambiente 2019, 26, e20170594. [Google Scholar] [CrossRef]
- Freitas, C.S.A.; Maciel, L.F.; Corrêa dos Santos, R.A.; Costa, O.M.M.M.; Maia, F.C.B.; Rabelo, R.S.; Franco, H.C.J.; Alves, E.; Consonni, S.R.; Freitas, R.O.; et al. Bacterial Volatile Organic Compounds Induce Adverse Ultrastructural Changes and DNA Damage to the Sugarcane Pathogenic Fungus Thielaviopsis Ethacetica. Environ. Microbiol. 2022, 24, 1430–1453. [Google Scholar] [CrossRef]
- Dandurishvili, N.; Toklikishvili, N.; Ovadis, M.; Eliashvili, P.; Giorgobiani, N.; Keshelava, R.; Tediashvili, M.; Vainstein, A.; Khmel, I.; Szegedi, E.; et al. Broad-Range Antagonistic Rhizobacteria Pseudomonas Fluorescens and Serratia Plymuthica Suppress Agrobacterium Crown Gall Tumours on Tomato Plants: Biocontrol of Agrobacterium by Bacterial Antagonists. J. Appl. Microbiol. 2011, 110, 341–352. [Google Scholar] [CrossRef]
- Jampilek, J. Potential of Agricultural Fungicides for Antifungal Drug Discovery. Expert Opin. Drug Discov. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Mazu, T.K.; Bricker, B.A.; Flores-Rozas, H.; Ablordeppey, S.Y. The Mechanistic Targets of Antifungal Agents: An Overview. Mini Rev. Med. Chem. 2016, 16, 555–578. [Google Scholar] [CrossRef]
- Farag, M.A.; Ryu, C.-M.; Sumner, L.W.; Paré, P.W. GC–MS SPME Profiling of Rhizobacterial Volatiles Reveals Prospective Inducers of Growth Promotion and Induced Systemic Resistance in Plants. Phytochemistry 2006, 67, 2262–2268. [Google Scholar] [CrossRef]
- Park, Y.-S.; Dutta, S.; Ann, M.; Raaijmakers, J.M.; Park, K. Promotion of Plant Growth by Pseudomonas Fluorescens Strain SS101 via Novel Volatile Organic Compounds. Biochem. Biophys. Res. Commun. 2015, 461, 361–365. [Google Scholar] [CrossRef]
- Yasmin, H.; Rashid, U.; Hassan, M.N.; Nosheen, A.; Naz, R.; Ilyas, N.; Sajjad, M.; Azmat, A.; Alyemeni, M.N. Volatile Organic Compounds Produced by Pseudomonas pseudoalcaligenes Alleviated drought Stress by Modulating Defense System in Maize (Zea mays L.). Physiol. Plant. 2021, 172, 896–911. [Google Scholar] [CrossRef]
- Che, J.; Liu, B.; Liu, G.; Chen, Q.; Lan, J. Volatile Organic Compounds Produced by Lysinibacillus sp. FJAT-4748 Possess Antifungal Activity against Colletotrichum Acutatum. Biocontrol Sci. Technol. 2017, 27, 1349–1362. [Google Scholar] [CrossRef]
- Fernando, W.G.D.; Ramarathnam, R.; Krishnamoorthy, A.S.; Savchuk, S.C. Identification and Use of Potential Bacterial Organic Antifungal Volatiles in Biocontrol. Soil Biol. Biochem. 2005, 37, 955–964. [Google Scholar] [CrossRef]
- Syed-Ab-Rahman, S.F.; Carvalhais, L.C.; Chua, E.; Xiao, Y.; Wass, T.J.; Schenk, P.M. Identification of Soil Bacterial Isolates Suppressing Different Phytophthora spp. and Promoting Plant Growth. Front. Plant Sci. 2018, 9, 1502. [Google Scholar] [CrossRef]
- Hunziker, L.; Bönisch, D.; Groenhagen, U.; Bailly, A.; Schulz, S.; Weisskopf, L. Pseudomonas Strains Naturally Associated with Potato Plants Produce Volatiles with High Potential for Inhibition of Phytophthora Infestans. Appl. Environ. Microbiol. 2015, 81, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Lo Cantore, P.; Giorgio, A.; Iacobellis, N.S. Bioactivity of Volatile Organic Compounds Produced by Pseudomonas Tolaasii. Front. Microbiol. 2015, 6, 156562. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, J.; Li, C.; Ma, Y. Antifungal and Plant Growth Promotion Activity of Volatile Organic Compounds Produced by Bacillus Amyloliquefaciens. MicrobiologyOpen 2019, 8, e00813. [Google Scholar] [CrossRef]
- Garbeva, P.; Hordijk, C.; Gerards, S.; de Boer, W. Volatiles Produced by the Mycophagous Soil Bacterium Collimonas. FEMS Microbiol. Ecol. 2014, 87, 639–649. [Google Scholar] [CrossRef]
- Garbeva, P.; Hordijk, C.; Gerards, S.; de Boer, W. Volatile-Mediated Interactions between Phylogenetically Different Soil Bacteria. Front. Microbiol. 2014, 5, 91242. [Google Scholar] [CrossRef]
- Ali, Q.; Khan, A.R.; Tao, S.; Rajer, F.U.; Ayaz, M.; Abro, M.A.; Gu, Q.; Wu, H.; Kuptsov, V.; Kolomiets, E.; et al. Broad-spectrum Antagonistic Potential of Bacillus spp. Volatiles against Rhizoctonia solani and Xanthomonas oryzae Pv. Oryzae. Physiol. Plant. 2023, 175, e14087. [Google Scholar] [CrossRef]
- Meldau, D.G.; Meldau, S.; Hoang, L.H.; Underberg, S.; Wunsche, H.; Baldwin, I.T. Dimethyl Disulfide Produced by the Naturally Associated Bacterium Bacillus sp. B55 Promotes Nicotiana attenuata Growth by Enhancing Sulfur Nutrition. Plant Cell 2013, 25, 2731–2747. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhou, J.; Tian, R.; Liu, Y. Microbial Volatile Organic Compounds: Antifungal Mechanisms, Applications, and Challenges. Front. Microbiol. 2022, 13, 922450. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ning, P.; Zheng, L.; Huang, J.; Li, G.; Hsiang, T. Effects of Volatile Substances of Streptomyces Globisporus JK-1 on Control of Botrytis Cinerea on Tomato Fruit. Biol. Control 2012, 61, 113–120. [Google Scholar] [CrossRef]
- Ebadzadsahrai, G.; Higgins Keppler, E.A.; Soby, S.D.; Bean, H.D. Inhibition of Fungal Growth and Induction of a Novel Volatilome in Response to Chromobacterium Vaccinii Volatile Organic Compounds. Front. Microbiol. 2020, 11, 1035. [Google Scholar] [CrossRef]
- Avis, T.J.; Bélanger, R.R. Specificity and Mode of Action of the Antifungal Fatty Acid Cis-9-Heptadecenoic Acid Produced by Pseudozyma Flocculosa. Appl. Environ. Microbiol. 2001, 67, 956–960. [Google Scholar] [CrossRef]
- Bergsson, G.; Arnfinnsson, J.; Steingrímsson, Ó.; Thormar, H. In Vitro Killing of Candida albicans by Fatty Acids and Monoglycerides. Antimicrob Agents Chemother 2001, 45, 3209–3212. [Google Scholar] [CrossRef]
- Zhang, H.; Godana, E.A.; Sui, Y.; Yang, Q.; Zhang, X.; Zhao, L. Biological Control as an Alternative to Synthetic Fungicides for the Management of Grey and Blue Mould Diseases of Table Grapes: A Review. Crit. Rev. Microbiol. 2020, 46, 450–462. [Google Scholar] [CrossRef]
- Stevens, S.; Hofmeyr, J.-H. Effects of Ethanol, Octanoic and Decanoic Acids on Fermentation and the Passive Influx of Protons through the Plasma Membrane of Saccharomyces Cerevisiae. Appl. Microbiol. Biotechnol. 1993, 38, 656–663. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabiço, F.; Borelli, T.C.; Alnoch, R.C.; Polizeli, M.d.L.T.d.M.; da Silva, R.R.; Silva-Rocha, R.; Guazzaroni, M.-E. Novel Pseudomonas Species Prevent the Growth of the Phytopathogenic Fungus Aspergillus flavus. BioTech 2024, 13, 8. https://doi.org/10.3390/biotech13020008
Rabiço F, Borelli TC, Alnoch RC, Polizeli MdLTdM, da Silva RR, Silva-Rocha R, Guazzaroni M-E. Novel Pseudomonas Species Prevent the Growth of the Phytopathogenic Fungus Aspergillus flavus. BioTech. 2024; 13(2):8. https://doi.org/10.3390/biotech13020008
Chicago/Turabian StyleRabiço, Franciene, Tiago Cabral Borelli, Robson Carlos Alnoch, Maria de Lourdes Teixeira de Moraes Polizeli, Ricardo R. da Silva, Rafael Silva-Rocha, and María-Eugenia Guazzaroni. 2024. "Novel Pseudomonas Species Prevent the Growth of the Phytopathogenic Fungus Aspergillus flavus" BioTech 13, no. 2: 8. https://doi.org/10.3390/biotech13020008




