Feature Papers in Biological Membrane Functions
Share This Topical Collection
Editors
 Dr. Katia Cortese
Dr. Katia Cortese
 Dr. Katia Cortese
Dr. Katia Cortese
E-Mail
Website
Collection Editor
Department of Experimental Medicine (DIMES), Cellular Electron Microscopy Lab., University of Genoa, 16132 Genova, Italy
Interests: breast cancers; targeted therapies; cell biology; endocytosis and trafficking; autophagy; electron microscopy; light microscopy; HER2/ERBB2 receptor; lysosomes
Special Issues, Collections and Topics in MDPI journals
 Dr. Jean Chemin
Dr. Jean Chemin
 Dr. Jean Chemin
Dr. Jean Chemin
E-Mail
Website
Collection Editor
Département de Neurosciences, Institut de Génomique Fonctionnelle, Centre National de la Recherche Scientifique, Unité Mixte de Recherche (UMR) 5203, Universités de Montpellier, 34094 Montpellier, France
Interests: ion channels; calcium channels; signal transduction; bioactive lipids; cannabinoids; sensory neurons; pain perception
 Prof. Dr. Byung Joo Kim
Prof. Dr. Byung Joo Kim
 Prof. Dr. Byung Joo Kim
Prof. Dr. Byung Joo Kim
E-Mail
Website
Collection Editor
Division of Longevity and Biofunctional Medicine, School of Korean Medicine, Pusan National University, Yangsan 50612, Republic of Korea
Interests: ion channel; smooth muscle; gastrointestinal motility; apoptosis; interstitial cells of cajal
 Prof. Dr. Rebecca J. Green
Prof. Dr. Rebecca J. Green
 Prof. Dr. Rebecca J. Green
Prof. Dr. Rebecca J. Green
E-Mail
Website
Collection Editor
Reading School of Pharmacy, University of Reading, Reading RG6 6DZ, UK
Interests: interactions with lipid membranes; antimicrobial peptides
Topical Collection Information
Dear Colleagues,
The field of biological membrane functions encompasses a diverse array of processes that are crucial for the functioning of living organisms. This topical collection of papers delves into the intricate mechanisms and key players involved in various aspects of membrane biology in health and disease. From ion channels to membrane fusion, each topic sheds light on fundamental processes that govern cellular dynamics and signal transduction. The collection aims to provide a comprehensive overview of the latest research findings and perspectives in this rapidly evolving field.
We look forward to receiving your outstanding contributions.
Dr. Katia Cortese
Dr. Jean Chemin
Prof. Dr. Byung Joo Kim
Prof. Dr. Rebecca J. Green
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Membranes is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- ion channels
- membrane transporters
- membrane permeability
- membrane trafficking processes
- signal transduction
- endocytosis
- autophagy
- membrane receptors
- protein–membrane interactions
- membrane fusion
- membrane remodeling
Published Papers (2 papers)
Open AccessArticle
An Untargeted Metabolomics Strategy to Identify Substrates of Known and Orphan E. coli Transporters
by
Mohammad S. Radi, Lachlan J. Munro, Daniela Rago and Douglas B. Kell
Viewed by 871
Abstract
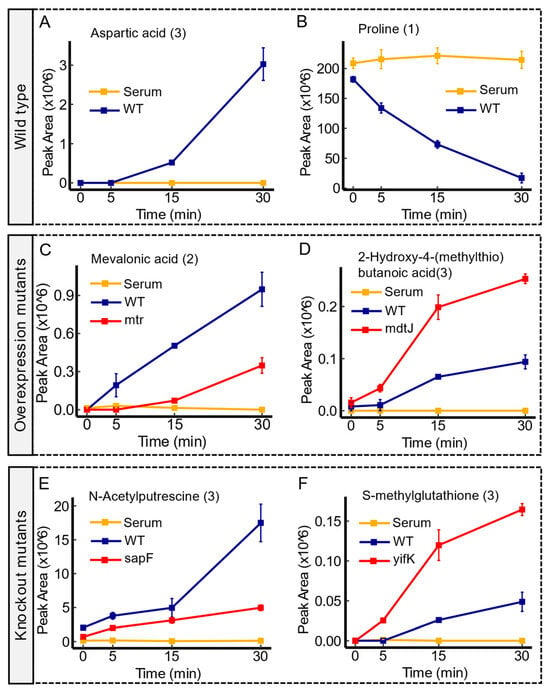
Transport systems play a pivotal role in bacterial physiology and represent potential targets for medical and biotechnological applications. However, even in well-studied organisms like
Escherichia coli, a notable proportion of transporters, exceeding as many as 30%, remain classified as orphans due to
[...] Read more.
Transport systems play a pivotal role in bacterial physiology and represent potential targets for medical and biotechnological applications. However, even in well-studied organisms like
Escherichia coli, a notable proportion of transporters, exceeding as many as 30%, remain classified as orphans due to their lack of known substrates. This study leveraged high-resolution LC-MS-based untargeted metabolomics to identify candidate substrates for these orphan transporters. Human serum, including a diverse array of biologically relevant molecules, served as an unbiased source for substrate exposure. The analysis encompassed 26 paired transporter mutant contrasts (i.e., knockout vs. overexpression), compared with the wild type, revealing distinct patterns of substrate uptake and excretion across various mutants. The convergence of candidate substrates across mutant scenarios provided robust validation, shedding light on novel transporter-substrate relationships, including those involving
yeaV,
hsrA,
ydjE, and
yddA. Furthermore, several substrates were contingent upon the specific mutants employed. This investigation underscores the utility of untargeted metabolomics for substrate identification in the absence of prior knowledge and lays the groundwork for subsequent validation experiments, holding significant implications for both medical and biotechnological advancements.
Full article
►▼
Show Figures
Open AccessArticle
Expression, Function and Trafficking of the Human ABCG2 Multidrug Transporter Containing Mutations in an Unstructured Cytoplasmic Loop
by
Orsolya Mózner, Boglárka Zámbó, Zsuzsa Bartos, Anna Gergely, Kata Sára Szabó, Bálint Jezsó, Ágnes Telbisz, György Várady, László Homolya, Tamás Hegedűs and Balázs Sarkadi
Viewed by 1411
Abstract
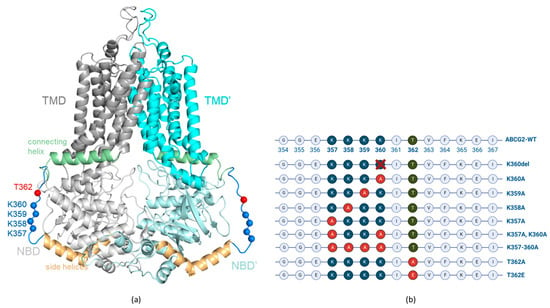
The human ABCG2 multidrug transporter plays a crucial role in the absorption and excretion of xeno- and endobiotics, contributes to cancer drug resistance and the development of gout. In this work, we have analyzed the effects of selected variants, residing in a structurally
[...] Read more.
The human ABCG2 multidrug transporter plays a crucial role in the absorption and excretion of xeno- and endobiotics, contributes to cancer drug resistance and the development of gout. In this work, we have analyzed the effects of selected variants, residing in a structurally unresolved cytoplasmic region (a.a. 354–367) of ABCG2 on the function and trafficking of this protein. A cluster of four lysines (K357–360) and the phosphorylation of a threonine (T362) residue in this region have been previously suggested to significantly affect the cellular fate of ABCG2. Here, we report that the naturally occurring K360del variant in human cells increased ABCG2 plasma membrane expression and accelerated cellular trafficking. The variable alanine replacements of the neighboring lysines had no significant effect on transport function, and the apical localization of ABCG2 in polarized cells has not been altered by any of these mutations. Moreover, in contrast to previous reports, we found that the phosphorylation-incompetent T362A, or the phosphorylation-mimicking T362E variants in this loop had no measurable effects on the function or expression of ABCG2. Molecular dynamics simulations indicated an increased mobility of the mutant variants with no major effects on the core structure of the protein. These results may help to decipher the potential role of this unstructured region within this transporter.
Full article
►▼
Show Figures