Genetic Modifiers at the Crossroads of Personalised Medicine for Haemoglobinopathies
Abstract
1. Introduction
2. Methods
2.1. Data Collection and Preprocessing
2.2. Development of an Evidence-Based Approach for Gene Ranking
2.3. Functional Enrichment Analysis
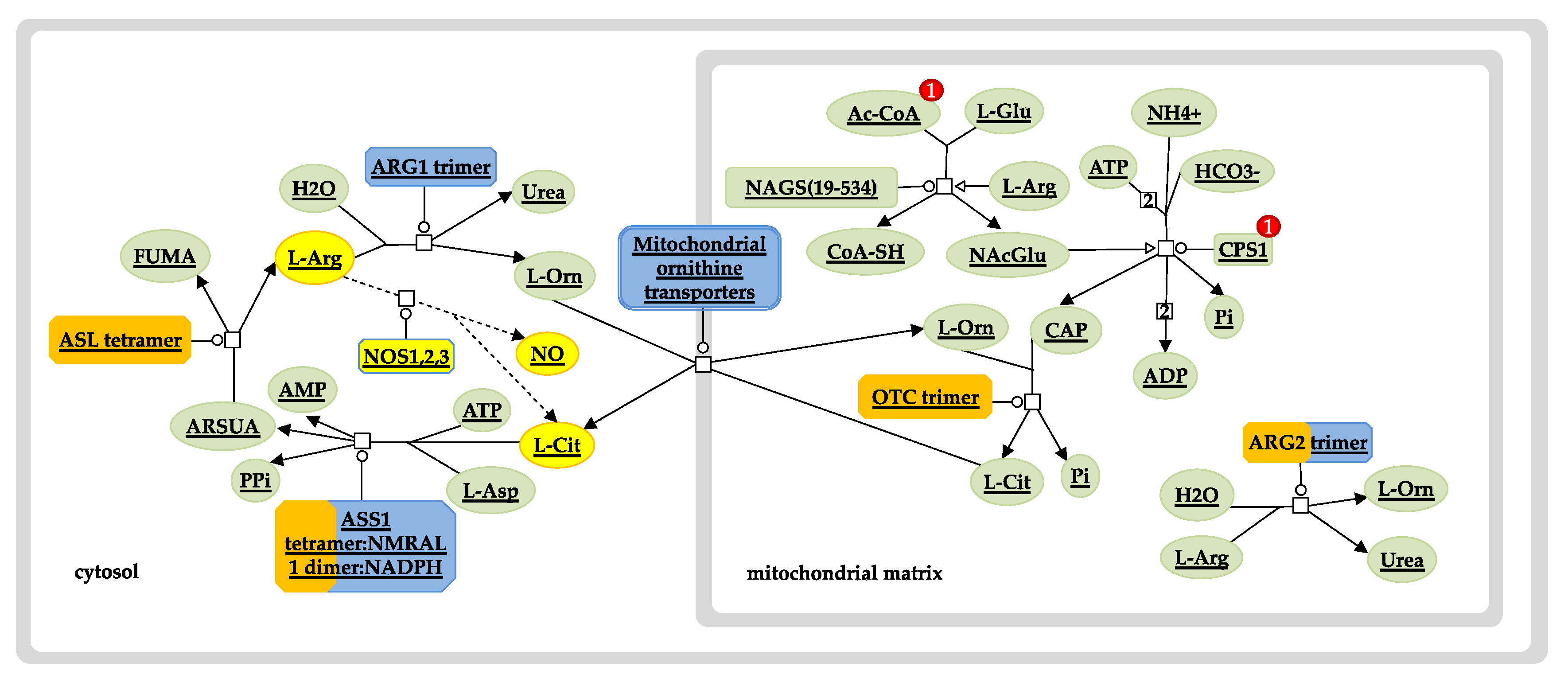
3. Results and Discussion
3.1. Exploratory Analysis of Modifier Gene Lists
3.2. Evidence-Based Gene Ranking
3.3. Functional Enrichment Analysis for Selected Phenotypes
3.3.1. Hb F Levels and Hb F Response to Hydroxyurea
3.3.2. Response to Iron Chelators
3.3.3. Stroke
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Modell, B.; Darlison, M. Global epidemiology of haemoglobin disorders and derived service indicators. Bull. World Health Organ. 2008, 86, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Weatherall, D.J.; Clegg, J.B. Inherited haemoglobin disorders: An increasing global health problem. Bull. World Health Organ. 2001, 79, 704–712. [Google Scholar] [PubMed]
- Piel, F.B.; Tatem, A.J.; Huang, Z.; Gupta, S.; Williams, T.N.; Weatherall, D.J. Global migration and the changing distribution of sickle haemoglobin: A quantitative study of temporal trends between 1960 and 2000. Lancet Glob. Health 2014, 2, e80–e89. [Google Scholar] [CrossRef]
- Henderson, S.; Timbs, A.; McCarthy, J.; Gallienne, A.; Van Mourik, M.; Masters, G.; May, A.; Khalil, M.S.M.; Schuh, A.; Old, J. Incidence of haemoglobinopathies in various populations—The impact of immigration. Clin. Biochem. 2009, 42, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Kountouris, P.; Lederer, C.W.; Fanis, P.; Feleki, X.; Old, J.; Kleanthous, M. IthaGenes: An Interactive Database for Haemoglobin Variations and Epidemiology. PLoS ONE 2014, 9, e103020. [Google Scholar] [CrossRef] [PubMed]
- Kountouris, P.; Stephanou, C.; Bento, C.; Fanis, P.; Elion, J.; Ramesar, R.S.; Zilfalil, B.A.; Robinson, H.M.; Traeger-Synodinos, J.; Human Variome Project Global Globin 2020 Challenge; et al. ITHANET: Information and database community portal for haemoglobinopathies. bioRxiv 2017, 209361. [Google Scholar] [CrossRef]
- Galanello, R.; Origa, R. Beta-thalassemia. Orphanet J. Rare Dis. 2010, 5, 11. [Google Scholar] [CrossRef]
- Rees, D.C.; Williams, T.N.; Gladwin, M.T. Sickle-cell disease. Lancet 2010, 376, 2018–2031. [Google Scholar] [CrossRef]
- Sripichai, O.; Munkongdee, T.; Kumkhaek, C.; Svasti, S.; Winichagoon, P.; Fucharoen, S. Coinheritance of the different copy numbers of alpha-globin gene modifies severity of beta-thalassemia/Hb E disease. Ann. Hematol. 2008, 87, 375–379. [Google Scholar] [CrossRef]
- Higgs, D.R.; Aldridge, B.E.; Lamb, J.; Clegg, J.B.; Weatherall, D.J.; Hayes, R.J.; Grandison, Y.; Lowrie, Y.; Mason, K.P.; Serjeant, B.E.; et al. The Interaction of Alpha-Thalassemia and Homozygous Sickle-Cell Disease. N. Engl. J. Med. 1982, 306, 1441–1446. [Google Scholar] [CrossRef]
- Thein, S.L.; Menzel, S. Discovering the genetics underlying foetal haemoglobin production in adults. Br. J. Haematol. 2009, 145, 455–467. [Google Scholar] [CrossRef]
- Powars, D.R.; Weiss, J.N.; Chan, L.S.; Schroeder, W.A. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood 1984, 63, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Musallam, K.M.; Sankaran, V.G.; Cappellini, M.D.; Duca, L.; Nathan, D.G.; Taher, A.T. Fetal hemoglobin levels and morbidity in untransfused patients with β-thalassemia intermedia. Blood 2012, 119, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Thein, S.L.; Menzel, S.; Lathrop, M.; Garner, C. Control of fetal hemoglobin: New insights emerging from genomics and clinical implications. Hum. Mol. Genet. 2009, 18, R216–R223. [Google Scholar] [CrossRef] [PubMed]
- Galarneau, G.; Palmer, C.D.; Sankaran, V.G.; Orkin, S.H.; Hirschhorn, J.N.; Lettre, G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet. 2010, 42, 1049–1051. [Google Scholar] [CrossRef]
- Uda, M.; Galanello, R.; Sanna, S.; Lettre, G.; Sankaran, V.G.; Chen, W.; Usala, G.; Busonero, F.; Maschio, A.; Albai, G.; et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc. Natl. Acad. Sci. USA 2008, 105, 1620–1625. [Google Scholar] [CrossRef]
- Mtatiro, S.N.; Singh, T.; Rooks, H.; Mgaya, J.; Mariki, H.; Soka, D.; Mmbando, B.; Msaki, E.; Kolder, I.; Thein, S.L.; et al. Genome Wide Association Study of Fetal Hemoglobin in Sickle Cell Anemia in Tanzania. PLoS ONE 2014, 9, e111464. [Google Scholar] [CrossRef]
- Liu, L.; Pertsemlidis, A.; Ding, L.-H.; Story, M.D.; Steinberg, M.H.; Sebastiani, P.; Hoppe, C.; Ballas, S.K.; Pace, B.S. Original Research: A case-control genome-wide association study identifies genetic modifiers of fetal hemoglobin in sickle cell disease. Exp. Biol. Med. 2016, 241, 706–718. [Google Scholar] [CrossRef]
- Schaefer, B.A.; Flanagan, J.M.; Alvarez, O.A.; Nelson, S.C.; Aygun, B.; Nottage, K.A.; George, A.; Roberts, C.W.; Piccone, C.M.; Howard, T.A.; et al. Genetic Modifiers of White Blood Cell Count, Albuminuria and Glomerular Filtration Rate in Children with Sickle Cell Anemia. PLoS ONE 2016, 11, e0164364. [Google Scholar] [CrossRef]
- Aguiar, L.; Matos, A.; Gil, Â.; Afonso, C.; Almeida, S.; Braga, L.; Lavinha, J.; Kjollerstrom, P.; Faustino, P.; Bicho, M.; et al. Sickle cell anemia—Nitric oxide related genetic modifiers of hematological and biochemical parameters. Clin. Hemorheol. Microcirc. 2016, 64, 957–963. [Google Scholar] [CrossRef]
- Flanagan, J.M.; Frohlich, D.M.; Howard, T.A.; Schultz, W.H.; Driscoll, C.; Nagasubramanian, R.; Mortier, N.A.; Kimble, A.C.; Aygun, B.; Adams, R.J.; et al. Genetic predictors for stroke in children with sickle cell anemia. Blood 2011, 117, 6681–6684. [Google Scholar] [CrossRef]
- Desai, A.A.; Zhou, T.; Ahmad, H.; Zhang, W.; Mu, W.; Trevino, S.; Wade, M.S.; Raghavachari, N.; Kato, G.J.; Peters-Lawrence, M.H.; et al. A Novel Molecular Signature for Elevated Tricuspid Regurgitation Velocity in Sickle Cell Disease. Am. J. Respir. Crit. Care Med. 2012, 186, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.A.; Novelli, E.M.; Isenberg, J.S.; Garrett, M.E.; Chu, Y.; Soldano, K.; Ataga, K.I.; Telen, M.J.; Ashley-Koch, A.; Gladwin, M.T.; et al. Thrombospondin-1 Gene Polymorphism is Associated with Estimated Pulmonary Artery Pressure in Patients with Sickle Cell Anemia. Am. J. Hematol. 2017, 92, E31–E34. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Gibson, J.S. Biomarkers in sickle cell disease. Br. J. Haematol. 2012, 156, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A. Biomarkers in drug discovery and development. Eur. Pharm. Rev. 2010, 5, 19–25. [Google Scholar]
- Kalpatthi, R.; Novelli, E.M. Measuring success: Utility of biomarkers in sickle cell disease clinical trials and care. Hematology 2018, 2018, 482–492. [Google Scholar] [CrossRef]
- Thein, S.L. Genetic modifiers of the beta-haemoglobinopathies. Br. J. Haematol. 2008, 141, 357–366. [Google Scholar] [CrossRef]
- Steinberg, M.H.; Sebastiani, P. Genetic modifiers of sickle cell disease. Am. J. Hematol. 2012, 87, 795–803. [Google Scholar] [CrossRef]
- Driss, A.; Asare, K.O.; Hibbert, J.M.; Gee, B.E.; Adamkiewicz, T.V.; Stiles, J.K. Sickle Cell Disease in the Post Genomic Era: A Monogenic Disease with a Polygenic Phenotype. Genom. Insights 2009, 2, 23–48. [Google Scholar] [CrossRef]
- Fertrin, K.Y.; Costa, F.F. Genomic polymorphisms in sickle cell disease: Implications for clinical diversity and treatment: Expert Review of Hematology: Vol 3, No 4. Expert Rev. Hematol. 2010, 3, 443–458. [Google Scholar] [CrossRef]
- Giardine, B.; Borg, J.; Viennas, E.; Pavlidis, C.; Moradkhani, K.; Joly, P.; Bartsakoulia, M.; Riemer, C.; Miller, W.; Tzimas, G.; et al. Updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Nucleic Acids Res. 2014, 42, D1063–D1069. [Google Scholar] [CrossRef]
- Sun, J.; Jia, P.; Fanous, A.H.; Webb, B.T.; van den Oord, E.J.C.G.; Chen, X.; Bukszar, J.; Kendler, K.S.; Zhao, Z. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases—Schizophrenia as a case. Bioinformatics 2009, 25, 2595–6602. [Google Scholar] [CrossRef]
- Larsen, E.; Menashe, I.; Ziats, M.N.; Pereanu, W.; Packer, A.; Banerjee-Basu, S. A systematic variant annotation approach for ranking genes associated with autism spectrum disorders. Mol. Autism 2016, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Li, J.; Shao, Q.; Chen, H.; Lin, Z.; Sun, Z.S.; Wu, J. EpilepsyGene: A genetic resource for genes and mutations related to epilepsy. Nucleic Acids Res. 2015, 43, D893–D899. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, J.; Rao, S.; Ritter, M.; Manor, L.C.; Backer, R.; Cao, H.; Cheng, Z.; Liu, S.; Liu, Y.; et al. An Integrative Computational Approach to Evaluate Genetic Markers for Bipolar Disorder. Sci. Rep. 2017, 7, 6745. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Kuo, P.-H.; Riley, B.P.; Kendler, K.S.; Zhao, Z. Candidate genes for schizophrenia: A survey of association studies and gene ranking. Am. J. Med. Genet. Part B 2008, 147, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Mantzaris, D.; Georgitsi, M.; Drineas, P.; Paschou, P. Variant Ranker: A web-tool to rank genomic data according to functional significance. BMC Bioinform. 2017, 18, 341. [Google Scholar] [CrossRef]
- Köhler, S.; Doelken, S.C.; Mungall, C.J.; Bauer, S.; Firth, H.V.; Bailleul-Forestier, I.; Black, G.C.M.; Brown, D.L.; Brudno, M.; Campbell, J.; et al. The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014, 42, D966–D974. [Google Scholar] [CrossRef]
- Robinson, P.N.; Köhler, S.; Bauer, S.; Seelow, D.; Horn, D.; Mundlos, S. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am. J. Hum. Genet. 2008, 83, 610–615. [Google Scholar] [CrossRef]
- Strande, N.T.; Riggs, E.R.; Buchanan, A.H.; Ceyhan-Birsoy, O.; DiStefano, M.; Dwight, S.S.; Goldstein, J.; Ghosh, R.; Seifert, B.A.; Sneddon, T.P.; et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 2017, 100, 895–906. [Google Scholar] [CrossRef]
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A tool for interactive Gene Ontology analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2019. Nucleic Acids Res. 2019, 47, D948–D954. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exloration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef]
- MacArthur, D.G.; Manolio, T.A.; Dimmock, D.P.; Rehm, H.L.; Shendure, J.; Abecasis, G.R.; Adams, D.R.; Altman, R.B.; Antonarakis, S.E.; Ashley, E.A.; et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–476. [Google Scholar] [CrossRef]
- Ah, Y.-M.; Kim, Y.-M.; Kim, M.-J.; Choi, Y.H.; Park, K.-H.; Son, I.-J.; Kim, S.G. Drug-induced Hyperbilirubinemia and the Clinical Influencing Factors. Drug Metab. Rev. 2008, 40, 511–537. [Google Scholar] [CrossRef]
- Lettre, G.; Sankaran, V.G.; Bezerra, M.A.C.; Araújo, A.S.; Uda, M.; Sanna, S.; Cao, A.; Schlessinger, D.; Costa, F.F.; Hirschhorn, J.N.; et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and β-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. USA 2008, 105, 11869–11874. [Google Scholar] [CrossRef]
- Borg, J.; Papadopoulos, P.; Georgitsi, M.; Gutiérrez, L.; Grech, G.; Fanis, P.; Phylactides, M.; Verkerk, A.J.M.H.; van der Spek, P.J.; Scerri, C.A.; et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes Hereditary Persistence of Fetal Hemoglobin. Nat. Genet. 2010, 42, 801–805. [Google Scholar] [CrossRef]
- Menzel, S.; Thein, S.L. Genetic Modifiers of Fetal Haemoglobin in Sickle Cell Disease. Mol. Diagn. Ther. 2019, 23, 235–244. [Google Scholar] [CrossRef]
- Ware, R.E. How I use hydroxyurea to treat young patients with sickle cell anemia. Blood 2010, 115, 5300–5311. [Google Scholar] [CrossRef]
- Thein, S.L. Genetic Basis and Genetic Modifiers of β-Thalassemia and Sickle Cell Disease. In Gene and Cell Therapies for Beta-Globinopathies; Malik, P., Tisdale, J., Eds.; Advances in Experimental Medicine and Biology; Springer New York: New York, NY, USA, 2017; pp. 27–57. ISBN 978-1-4939-7299-9. [Google Scholar]
- Pule, G.D.; Mowla, S.; Novitzky, N.; Wiysonge, C.S.; Wonkam, A. A Systematic Review of Known Mechanisms of Hydroxyurea-induced Foetal Haemoglobin for Treatment of Sickle Cell Disease. Expert Rev. Hematol. 2015, 8, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Ribeil, J.-A.; Arlet, J.-B.; Dussiot, M.; Cruz Moura, I.; Courtois, G.; Hermine, O. Ineffective Erythropoiesis in β-Thalassemia. Sci. World J. 2013, 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhou, S.; Kihm, A.J.; Katein, A.M.; Yu, X.; Gell, D.A.; Mackay, J.P.; Adachi, K.; Foster-Brown, L.; Louden, C.S.; et al. Loss of α-hemoglobin–stabilizing protein impairs erythropoiesis and exacerbates β-thalassemia. J. Clin. Investig. 2004, 114, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, V.G.; Ludwig, L.S.; Sicinska, E.; Xu, J.; Bauer, D.E.; Eng, J.C.; Patterson, H.C.; Metcalf, R.A.; Natkunam, Y.; Orkin, S.H.; et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 2012, 26, 2075–2087. [Google Scholar] [CrossRef] [PubMed]
- Manwani, D.; Frenette, P.S. Vaso-occlusion in sickle cell disease: Pathophysiology and novel targeted therapies. Blood 2013, 122, 3892–3898. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Bakshi, N.; Krishnamurti, L. Acute Chest Syndrome in Children with Sickle Cell Disease. Pediatr. Allergy Immunol. Pulmonol. 2017, 30, 191–201. [Google Scholar] [CrossRef]
- Desai, P.C.; Ataga, K.I. The acute chest syndrome of sickle cell disease. Expert Opin. Pharmacother. 2013, 14, 991–999. [Google Scholar] [CrossRef]
- Dichgans, M. Genetics of ischaemic stroke. Lancet Neurol. 2007, 6, 149–161. [Google Scholar] [CrossRef]
- Steinberg, M.H.; Lu, Z.-H.; Barton, F.B.; Terrin, M.L.; Charache, S.; Dover, G.J. Fetal Hemoglobin in Sickle Cell Anemia: Determinants of Response to Hydroxyurea. Blood 1997, 89, 1078–1088. [Google Scholar] [CrossRef]
- Lebensburger, J.D.; Pestina, T.I.; Ware, R.E.; Boyd, K.L.; Persons, D.A. Hydroxyurea therapy requires HbF induction for clinical benefit in a sickle cell mouse model. Haematologica 2010, 95, 1599–1603. [Google Scholar] [CrossRef]
- King, S.B. A role for nitric oxide in hydroxyurea-mediated fetal hemoglobin induction. J. Clin. Investig. 2003, 111, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Cokic, V.P.; Smith, R.D.; Beleslin-Cokic, B.B.; Njoroge, J.M.; Miller, J.L.; Gladwin, M.T.; Schechter, A.N. Hydroxyurea induces fetal hemoglobin by the nitric oxide—dependent activation of soluble guanylyl cyclase. J. Clin. Investig. 2003, 111, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Ikuta, T.; Ausenda, S.; Cappellini, M.D. Mechanism for fetal globin gene expression: Role of the soluble guanylate cyclase–cGMP-dependent protein kinase pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 1847–1852. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Morris, S.M. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Denninger, J.W.; Marletta, M.A. Guanylate cyclase and the ⋅NO/cGMP signaling pathway. Biochim. Biophys. Acta BBA - Bioenerg. 1999, 1411, 334–350. [Google Scholar] [CrossRef]
- Bhatta, S.S.; Wroblewski, K.E.; Agarwal, K.L.; Sit, L.; Cohen, E.E.W.; Seiwert, T.Y.; Karrison, T.; Bakris, G.L.; Ratain, M.J.; Vokes, E.E.; et al. Effects of Vascular Endothelial Growth Factor Signaling Inhibition on Human Erythropoiesis. Oncologist 2013, 18, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, A.C.; Licht, T.; Kumar, S.; Oladipupo, S.S.; Iyer, S.; Grunewald, M.; Keshet, E. VEGF expands erythropoiesis via hypoxia-independent induction of erythropoietin in noncanonical perivascular stromal cells. J. Exp. Med. 2019, 216, 215–230. [Google Scholar] [CrossRef]
- Drogat, B.; Kalucka, J.; Gutierrez, L.; Hammad, H.; Goossens, S.; Farhang Ghahremani, M.; Bartunkova, S.; Haigh, K.; Deswarte, K.; Nyabi, O.; et al. Vegf regulates embryonic erythroid development through Gata1 modulation. Blood 2010, 116, 2141–2151. [Google Scholar] [CrossRef]
- Fang, S.; Nurmi, H.; Heinolainen, K.; Chen, S.; Salminen, E.; Saharinen, P.; Mikkola, H.K.A.; Alitalo, K. Critical requirement of VEGF-C in transition to fetal erythropoiesis. Blood 2016, 128, 710–720. [Google Scholar] [CrossRef]
- Cao, A.; Moi, P.; Galanello, R. Recent advances in β-thalassemias. Pediatric Rep. 2011, 3, e17. [Google Scholar] [CrossRef]
- Galanello, R.; Campus, S.; Origa, R. Deferasirox: Pharmacokinetics and clinical experience. Expert Opin. Drug Metab. Toxicol. 2012, 8, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, N.F.; Brittenham, G.M.; McLaren, C.E.; Templeton, D.M.; Cameron, R.G.; McClelland, R.A.; Burt, A.D.; Fleming, K.A. Long-Term Safety and Effectiveness of Iron-Chelation Therapy with Deferiprone for Thalassemia Major. N. Engl. J. Med. 2009, 339, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Galanello, R. Deferiprone in the treatment of transfusion-dependent thalassemia: A review and perspective. Ther. Clin. Risk Manag. 2007, 3, 795–805. [Google Scholar] [PubMed]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Münzel, P.A.; Schmohl, S.; Heel, H.; Kälberer, K.; Bock-Hennig, B.S.; Bock, K.W. Induction of Human UDP Glucuronosyltransferases (UGT1A6, UGT1A9, and UGT2B7) by t-Butylhydroquinone and 2,3,7,8-Tetrachlorodibenzo-p-Dioxin in Caco-2 Cells. Drug Metab. Dispos. 1999, 27, 569–573. [Google Scholar]
- Nebert, D.W.; Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 2006, 6, 947–960. [Google Scholar] [CrossRef]
- Exjade-European Public Assessment Report; European Medicines Evaluation Agency: London, UK, 2006.
- Waldmeier, F.; Bruin, G.J.; Glaenzel, U.; Hazell, K.; Sechaud, R.; Warrington, S.; Porter, J.B. Pharmacokinetics, Metabolism, and Disposition of Deferasirox in β-Thalassemic Patients with Transfusion-Dependent Iron Overload Who Are at Pharmacokinetic Steady State. Drug Metab. Dispos. 2010, 38, 808–816. [Google Scholar] [CrossRef]
- Bruin, G.J.M.; Faller, T.; Wiegand, H.; Schweitzer, A.; Nick, H.; Schneider, J.; Boernsen, K.-O.; Waldmeier, F. Pharmacokinetics, Distribution, Metabolism, and Excretion of Deferasirox and Its Iron Complex in Rats. Drug Metab. Dispos. 2008, 36, 2523–2538. [Google Scholar] [CrossRef]
- Jemnitz, K.; Heredi-Szabo, K.; Janossy, J.; Ioja, E.; Vereczkey, L.; Krajcsi, P. ABCC2/Abcc2: A multispecific transporter with dominant excretory functions. Drug Metab. Rev. 2010, 42, 402–436. [Google Scholar] [CrossRef]
- Haverfield, E.V.; Weatherall, D.J.; Graber, A.Y.; Ramirez, J.; Ratain, M.J. Pharmacogenomics of Deferiprone Metabolism. Blood 2005, 106, 2703. [Google Scholar]
- Benoit-Biancamano, M.-O.; Connelly, J.; Villeneuve, L.; Caron, P.; Guillemette, C. Deferiprone Glucuronidation by Human Tissues and Recombinant UDP Glucuronosyltransferase 1A6: An in Vitro Investigation of Genetic and Splice Variants. Drug Metab. Dispos. 2009, 37, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, M.; Groothuis, G.M.M.; de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, S.J.; Collier, A.C. Pediatric Development of Glucuronidation: The Ontogeny of Hepatic UGT1A4. Drug Metab. Dispos. 2007, 35, 1587–1592. [Google Scholar] [CrossRef]
- Miyagi, S.J.; Milne, A.M.; Coughtrie, M.W.H.; Collier, A.C. Neonatal Development of Hepatic UGT1A9: Implications of Pediatric Pharmacokinetics. Drug Metab. Dispos. 2012, 40, 1321–1327. [Google Scholar] [CrossRef]
- Kassim, A.A.; Galadanci, N.A.; Pruthi, S.; DeBaun, M.R. How I treat and manage strokes in sickle cell disease. Blood 2015, 125, 3401–3410. [Google Scholar] [CrossRef]
- Ohene-Frempong, K.; Weiner, S.J.; Sleeper, L.A.; Miller, S.T.; Embury, S.; Moohr, J.W.; Wethers, D.L.; Pegelow, C.H.; Gill, F.M. Cerebrovascular Accidents in Sickle Cell Disease: Rates and Risk Factors. Blood 1998, 91, 288–294. [Google Scholar]
- Quinn, C.T. Sickle Cell Disease in Childhood. Pediatr. Clin. N. Am. 2013, 60, 1363–1381. [Google Scholar] [CrossRef]
- Driscoll, M.C.; Hurlet, A.; Styles, L.; McKie, V.; Files, B.; Olivieri, N.; Pegelow, C.; Berman, B.; Drachtman, R.; Patel, K.; et al. Stroke risk in siblings with sickle cell anemia. Blood 2003, 101, 2401–2404. [Google Scholar] [CrossRef]
- Martella, M.; Quaglia, N.; Frigo, A.C.; Basso, G.; Colombatti, R.; Sainati, L. Association between a combination of single nucleotide polymorphisms and large vessel cerebral vasculopathy in African children with sickle cell disease. Blood Cells. Mol. Dis. 2016, 61, 1–3. [Google Scholar] [CrossRef]
- Pan, W.; Kastin, A.J. Tumor necrosis factor and stroke: Role of the blood-brain barrier. Prog. Neurobiol. 2007, 83, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Henkler, F.; Scheurich, P. The TNF-receptor-associated factor family: Scaffold molecules for cytokine receptors, kinases and their regulators. Cell. Signal. 2001, 13, 389–400. [Google Scholar] [CrossRef]
- Wang, J.; Hu, Z.; Yang, S.; Liu, C.; Yang, H.; Wang, D.; Guo, F. Inflammatory cytokines and cells are potential markers for patients with cerebral apoplexy in intensive care unit. Exp. Ther. Med. 2018, 16, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Lambertsen, K.L.; Finsen, B.; Clausen, B.H. Post-stroke inflammation-target or tool for therapy? Acta Neuropathol. 2019, 137, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Jickling, G.C.; Sharp, F.R. Biomarker Panels in Ischemic Stroke. Stroke 2015, 46, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Moon, G.J.; Bang, O.Y. Biomarkers for Stroke. J. Stroke 2013, 15, 27–37. [Google Scholar] [CrossRef]
- Katan, M.; Elkind, M.S. The potential role of blood biomarkers in patients with ischemic stroke: An expert opinion. Clin. Transl. Neurosci. 2018, 2. [Google Scholar] [CrossRef]
- Fang, C.; Lou, B.; Zhou, J.; Zhong, R.; Wang, R.; Zang, X.; Shen, H.; Li, Y. Blood biomarkers in ischemic stroke: Role of biomarkers in differentiation of clinical phenotype. Eur. J. Inflamm. 2018, 16, 1–10. [Google Scholar] [CrossRef]
- Riordan, J.D.; Nadeau, J.H. From Peas to Disease: Modifier Genes, Network Resilience, and the Genetics of Health. Am. J. Hum. Genet. 2017, 101, 177–191. [Google Scholar] [CrossRef]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.A.; Hirschhorn, J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]







| Evidence | Type | Description | Points | |
| Association Score (AS) | Association study | p value | <0.05 | 0.5 |
| <0.001 | 1 | |||
| <0.00001 | 1.5 | |||
| Maximum Allowable Sum of Points for Association Score | 8 | |||
| Variant Score (VS) | Genetic variants | Number of variants | One point for each variant in every phenotype stored in IthaGenes. | 1 |
| Maximum Allowable Sum of Points for Variant Score | 20 | |||
| Experimental Score (ES) | Function | Biochemical Function | Functions are shared between gene products involved in the same disease phenotype. | 1 |
| Protein Interaction | Gene product interacts with proteins previously implicated in the disease phenotype. Gene defect disrupting protein interactions. | 1 | ||
| Expression | Gene is expressed in tissues relevant to the disease phenotype. Altered gene expression in patients. | 1 | ||
| Functional Alteration | Cells from affected individual | Function of gene product is altered in individuals/engineered cells with candidate mutations (altered expression levels, splicing or normal biochemical function). | 1.5 | |
| Engineered cells | 1.5 | |||
| Model Systems | Animal model | Introduction of the variant or an engineered gene product carrying the variant in a non-human animal model/cell-culture model displays the disease phenotype. | 2 | |
| Cell culture model system | 2 | |||
| Rescue | Rescue in non-human model organism | Addition of the wild-type gene product or specific knockdown of the variant allele can rescue the disease phenotype in a non-human model organism/cell-culture model/patient. | 2 | |
| Rescue in cell culture model | 2 | |||
| Rescue in patients | 2 | |||
| Maximum Allowable Sum of Points for Experimental Score | 6 | |||
| Phenotype ID | Phenotypic Term | HPO ID | Gene/Intergenic Region | IthaScore |
|---|---|---|---|---|
| 2 | Hb F levels | HP:0011904 | BCL11A | 0.8750 |
| 28 | Bilirubin levels | − | UGT1A1 | 0.4397 |
| 10 | F-cell numbers | − | HBS1L-MYB | 0.3169 |
| 5 | Ineffective erythropoiesis | HP:0010972 | AHSP, SOX6 | 0.3000 |
| 4 | Anaemia | HP:0001903 | CCND3 | 0.2938 |
| 11 | Globin gene regulation | − | SIRT1 | 0.2500 |
| 9 | Hb F response to hydroxyurea | − | HBG2 | 0.2188 |
| 7 | Focal segmental glomerulosclerosis | HP:0000097 | APOL1 | 0.2175 |
| 24 | Response to Hepatitis C treatment | − | IFNL3 | 0.2175 |
| 16 | Abnormal platelet count | HP:0011873 | HBS1L-MYB | 0.1997 |
| 29 | Gallstones | HP:0001081 | UGT1A1 | 0.1663 |
| 14 | Acute chest syndrome | − | EDN1 | 0.1450 |
| 23 | Vaso-occlusive crisis | − | HMOX1 | 0.1413 |
| 6 | Osteonecrosis/Avascular necrosis | HP:0010885 | KL | 0.1413 |
| 3 | Stroke | HP:0001297 | ENPP1 | 0.1350 |
| 22 | Increased serum ferritin | HP:0003281 | HFE | 0.1350 |
| 8 | Proteinuria | HP:0000093 | MYH9 | 0.1325 |
| 19 | Abnormal serum iron concentration | HP:0040130 | GDF15 | 0.1250 |
| 18 | Pain | HP:0012531 | GCH1 | 0.1184 |
| 17 | Left ventricular diastolic dysfunction | HP:0025168 | FUCA2 | 0.1100 |
| 1 | Abnormal red blood cell count | HP:0020058 | ABO, CCND3, PRKCE, PARP11-CCND2 | 0.1038 |
| 13 | Abnormal white blood cell count | HP:0011893 | CDK6, LY6G5C, PNPLA3, PSMD3-CSF3 | 0.1038 |
| 20 | Hyperuricemia | HP:0002149 | HBG1-HBG2 | 0.1038 |
| 21 | Abnormal hematocrit | HP:0031850 | HBS1L-MYB, PDGFRA-KIT | 0.1038 |
| 25 | Increased Hb A2 levels | HP:0045048 | LCRB | 0.1038 |
| 27 | Haemolytic anaemia | HP:0001878 | NPRL3 | 0.1038 |
| 26 | EPO levels | − | MAP2K6 | 0.1038 |
| 15 | Osteoporosis | HP:0000939 | COL1A1 | 0.1038 |
| 12 | Bacteremia | HP:0031864 | BMP6 | 0.1025 |
| 30 | Oxidative stress | HP:0025464 | FOXO3 | 0.1000 |
| 31 | Albuminuria | HP:0012592 | APOL1 | 0.0959 |
| 32 | Pulmonary arterial hypertension | HP:0002092 | NEDD4L | 0.0825 |
| 33 | RBC adhesion | − | ADCY6 | 0.0825 |
| 34 | Delayed menarche | HP:0012569 | NOS3 | 0.0825 |
| 35 | Red blood cell alloimmunisation | − | CD81 | 0.0825 |
| 36 | Reticulocytosis | HP:0001923 | NPRL3 | 0.0803 |
| 37 | Abnormal neutrophil cell number | HP:0011991 | NES | 0.0803 |
| 38 | Abnormal GFR | HP:0012212 | APOL1 | 0.0747 |
| 39 | Leg ulcers | − | SMAD7 | 0.0725 |
| 40 | Increased serum iron | HP:0003452 | HFE | 0.0725 |
| 41 | Cardiac iron load | − | GSTM1 | 0.0725 |
| 42 | Thromboembolism | HP:0001907 | PROC | 0.0613 |
| 43 | Response to Hydroxyurea | − | CD36 | 0.0600 |
| 44 | Priapism | HP:0200023 | AQP1, ITGAV, TGFBR3 | 0.0569 |
| 45 | Reticulocytopenia | HP:0001896 | BCL11A | 0.0569 |
| 46 | Recurrent respiratory infections | HP:0002205 | LGALS3 | 0.0513 |
| 47 | Increased lactate dehydrogenase activity | HP:0025435 | NOS3 | 0.0513 |
| 48 | Response to deferiprone | − | UGT1A6 | 0.0513 |
| 49 | Abnormal hepcidin level | HP:0031875 | TMPRSS6 | 0.0434 |
| 50 | Abnormal serum ferritin | HP:0040133 | GSTM1 | 0.0413 |
| 51 | Elevated transferrin saturation | HP:0012463 | HFE | 0.0413 |
| 52 | Decreased serum ferritin | HP:0012343 | TF, TFR2, TNF | 0.0413 |
| 53 | Abnormal circulating homocysteine concentration | HP:0010919 | MTHFR | 0.0413 |
| 54 | Morphine glucuronidation | − | UGT2B7 | 0.0413 |
| 55 | Increased liver iron level | HP:0012465 | HAMP | 0.0334 |
| 56 | Response to deferasirox | − | CYP1A2 | 0.0434 |
| 57 | Retinopathy | HP:0000488 | IL6, NOS3 | 0.0413 |
| 58 | Recurrent upper respiratory tract infections | HP:0002788 | NOS3 | 0.0413 |
| 59 | Recurrent Infections | HP:0002719 | CCL5, MPO, TLR2 | 0.0313 |
| Phenotype ID | Phenotypic Term | HPO ID | Gene/Intergenic Region | IthaScore |
|---|---|---|---|---|
| 2 | Hb F levels | HP:0011904 | BCL11A | 0.875 |
| 2 | Hb F levels | HP:0011904 | HBS1L-MYB | 0.825 |
| 2 | Hb F levels | HP:0011904 | KLF1 | 0.711 |
| 2 | Hb F levels | HP:0011904 | HBG2 | 0.600 |
| 2 | Hb F levels | HP:0011904 | HBE1 | 0.462 |
| 28 | Bilirubin levels | − | UGT1A1 | 0.440 |
| 2 | Hb F levels | HP:0011904 | HBG1 | 0.435 |
| 2 | Hb F levels | HP:0011904 | HBD-HBBP1 | 0.330 |
| 10 | F-cell numbers | − | HBS1L-MYB | 0.317 |
| 2 | Hb F levels | HP:0011904 | LCRB | 0.312 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stephanou, C.; Tamana, S.; Minaidou, A.; Papasavva, P.; Kleanthous, M.; Kountouris, P. Genetic Modifiers at the Crossroads of Personalised Medicine for Haemoglobinopathies. J. Clin. Med. 2019, 8, 1927. https://doi.org/10.3390/jcm8111927
Stephanou C, Tamana S, Minaidou A, Papasavva P, Kleanthous M, Kountouris P. Genetic Modifiers at the Crossroads of Personalised Medicine for Haemoglobinopathies. Journal of Clinical Medicine. 2019; 8(11):1927. https://doi.org/10.3390/jcm8111927
Chicago/Turabian StyleStephanou, Coralea, Stella Tamana, Anna Minaidou, Panayiota Papasavva, Marina Kleanthous, and Petros Kountouris. 2019. "Genetic Modifiers at the Crossroads of Personalised Medicine for Haemoglobinopathies" Journal of Clinical Medicine 8, no. 11: 1927. https://doi.org/10.3390/jcm8111927
APA StyleStephanou, C., Tamana, S., Minaidou, A., Papasavva, P., Kleanthous, M., & Kountouris, P. (2019). Genetic Modifiers at the Crossroads of Personalised Medicine for Haemoglobinopathies. Journal of Clinical Medicine, 8(11), 1927. https://doi.org/10.3390/jcm8111927







