Effects of Irregular Feeding on the Daily Fluctuations in mRNA Expression of the Neurosecretory Protein GL and Neurosecretory Protein GM Genes in the Mouse Hypothalamus
Abstract
:1. Introduction
2. Results
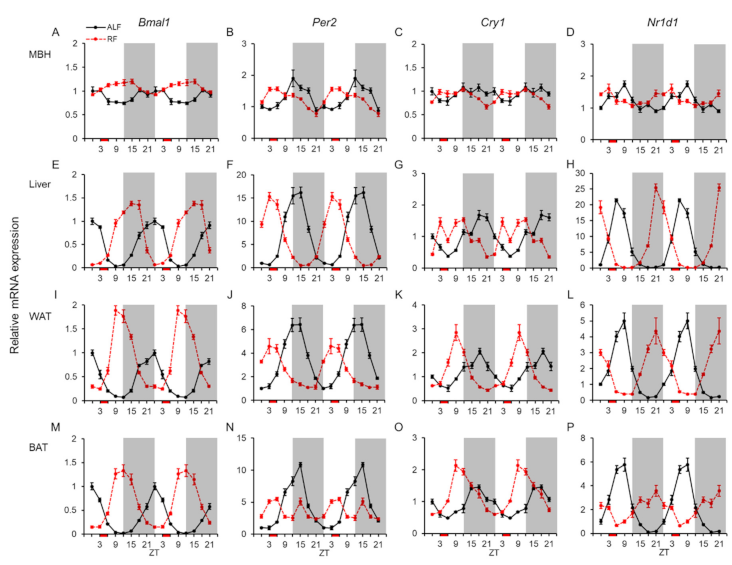
2.1. Daily mRNA Expression Profiles of Clock Genes in Central and Peripheral Tissues and Effects of the Feeding Times
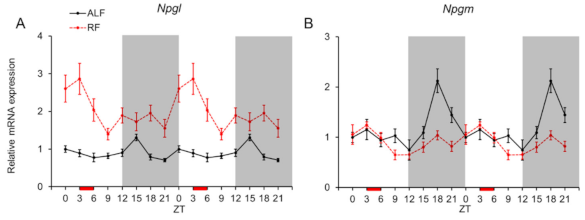
2.2. Daily mRNA Expression Profiles of Npgl and Npgm in MBH and Effects of the Feeding Times
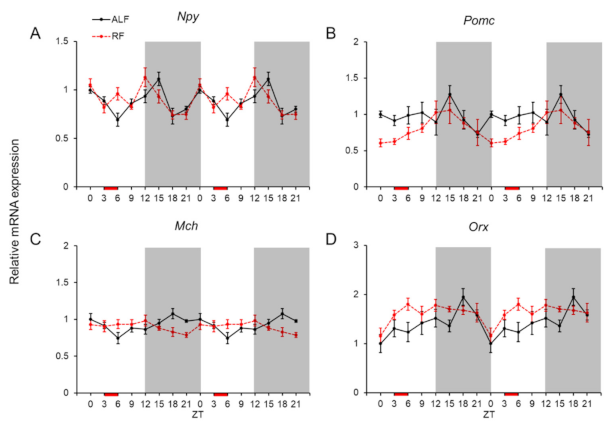
2.3. Daily mRNA Expression Profiles of Orexigenic and Anorexigenic Genes in MBH and Effects of the Feeding Times
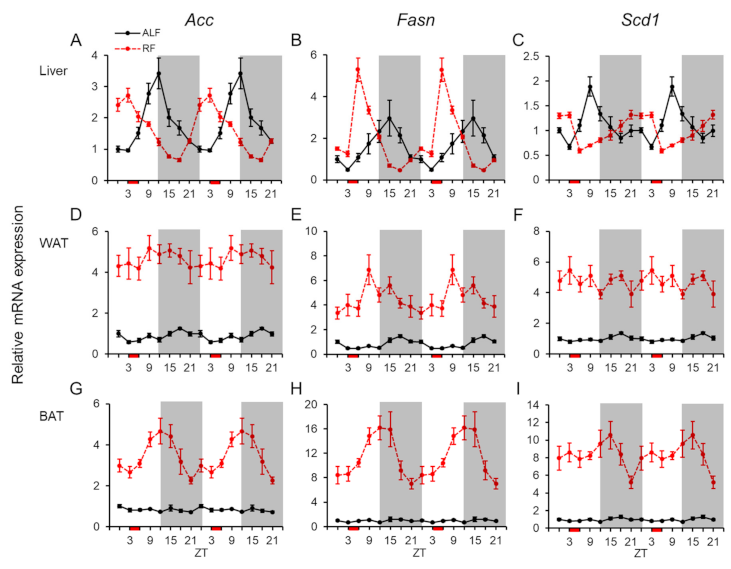
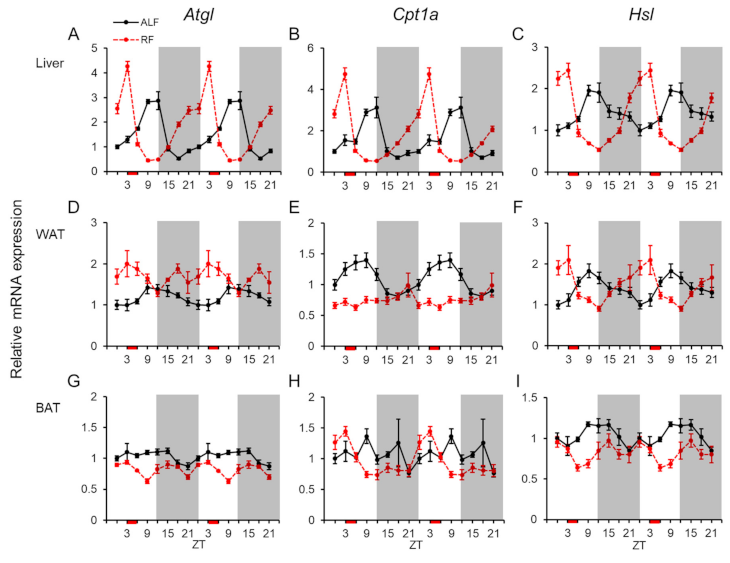
2.4. Daily mRNA Expression Profiles of Genes Related to Lipid Metabolism in Peripheral Tissues and Effects of the Feeding Times
2.5. Daily Profiles of Blood Serum Glucose and Insulin Levels and Effects of the Feeding Times
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Quantitative RT-PCR
4.3. Blood Tests
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Acetyl-CoA carboxylase |
| ACTB | Beta-actin |
| ALF | Ad libitum feeding |
| ATGL | Adipose triglyceride lipase |
| BAT | Brown adipose tissue |
| BMAL1 | Brain and muscle ARNT-like 1 |
| CPT1a | Carnitine palmitoyl transferase 1a |
| CRY1 | Cryptochrome 1 |
| DMH | Dorsomedial hypothalamus |
| FAA | Food anticipatory activity |
| FASN | Fatty acid synthase |
| HSL | Hormone-sensitive lipase |
| MBH | Mediobasal hypothalamus |
| MCH | Melanin-concentrating hormone |
| NPGL | Neurosecretory protein GL |
| NPGM | Neurosecretory protein GM |
| NPY | Neuropeptide Y |
| NR1D1 | Nuclear receptor subfamily 1 group D member 1 |
| ORX | Orexin |
| PER2 | Period 2 |
| POMC | Proopiomelanocortin |
| RPS18 | Ribosomal protein S18 |
| SCD1 | Stearoyl-CoA desaturase 1 |
| SCN | Suprachiasmatic nucleus |
| RF | Time-restricted feeding |
| WAT | White adipose tissue |
| ZT | Zeitgeber time |
References
- Kecklund, G.; Axelsson, J. Health consequences of shift work and insufficient sleep. Br. Med. J. 2016, 355, 5210. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.J.; Purvis, T.E.; Mistretta, J.; Scheer, F.A. Effects of the internal circadian system and circadian misalignment on glucose tolerance in chronic shift workers. J. Clin. Endocrinol. Metab. 2016, 101, 1066–1074. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U.; Albrecht, U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, M.S.; Young, M.E. The role of cell-specific circadian clocks in metabolism and disease. Obes. Rev. 2009, 10, 6–13. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Hastings, M.H.; Reddy, A.B.; Maywood, E.S. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003, 4, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Buhr, E.D.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Handb. Exp. Pharmacol. 2013, 217, 3–27. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Shearman, L.P.; Weaver, D.R.; Zylka, M.J.; de Vries, G.J.; Reppert, S.M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 1999, 96, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef]
- Bailey, S.M.; Udoh, U.S.; Young, M.E. Circadian regulation of metabolism. J. Endocrinol. 2014, 222, R75–R96. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, K.M.; Marcheva, B.; Kohsaka, A.; Bass, J. The clockwork of metabolism. Annu. Rev. Nutr. 2007, 27, 219–240. [Google Scholar] [CrossRef] [Green Version]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Triqueneaux, G.; Thenot, S.; Kakizawa, T.; Antoch, M.P.; Safi, R.; Takahashi, J.S.; Delaunay, F.; Laudet, V. The orphan receptor Rev-erb gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 2004, 33, 585–608. [Google Scholar] [CrossRef] [Green Version]
- Balsalobre, A. Clock genes in mammalian peripheral tissues. Cell Tissue Res. 2002, 309, 193–199. [Google Scholar] [CrossRef]
- Schibler, U.; Ripperger, J.; Brown, S.A. Peripheral circadian oscillators in mammals: Time and food. J. Biol. Rhythms 2003, 18, 250–260. [Google Scholar] [CrossRef]
- Shibata, S.; Tahara, Y.; Hirao, A. The adjustment and manipulation of biological rhythms by light, nutrition, and abused drugs. Adv. Drug Deliv. Rev. 2010, 62, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, R.; Wan, K.; Wakamatsu, H.; Aida, R.; Moriya, T.; Akiyama, M.; Shibata, S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells 2001, 6, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Stütz, A.M.; Staszkiewicz, J.; Ptitsyn, A.; Argyropoulos, G. Circadian expression of genes regulating food intake. Obesity 2007, 15, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Kalra, P.S.; Farmerie, W.G.; Kalra, S.P. Daily changes in hypothalamic gene expression of neuropeptide Y, galanin, proopiomelanocortin, and adipocyte leptin gene expression and secretion: Effects of food restriction. Endocrinology 1999, 140, 2868–2875. [Google Scholar] [CrossRef]
- Ramirez-Plascencia, O.D.; Saderi, N.; Escobar, C.; Salgado-Delgado, R.C. Feeding during the rest phase promotes circadian conflict in nuclei that control energy homeostasis and sleep-wake cycle in rats. Eur. J. Neurosci. 2017, 45, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Honma, S.; Honma, K. Effects of restricted daily feeding on neuropeptide Y release in the rat paraventricular nucleus. Am. J. Physiol. 1996, 270, 589–595. [Google Scholar] [CrossRef]
- Mukherji, A.; Kobiita, A.; Damara, M.; Misra, N.; Meziane, H.; Champy, M.F.; Chambon, P. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. USA 2015, 112, E6691–E6698. [Google Scholar] [CrossRef] [Green Version]
- Opperhuizen, A.L.; van Kerkhof, L.W.; Proper, K.I.; Rodenburg, W.; Kalsbeek, A. Rodent models to study the metabolic effects of shiftwork in humans. Front. Pharmacol. 2015, 6, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barclay, J.L.; Husse, J.; Bode, B.; Naujokat, N.; Meyer-Kovac, J.; Schmid, S.M.; Lehnert, H.; Oster, H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS ONE 2012, 7, e37150. [Google Scholar] [CrossRef] [Green Version]
- Opperhuizen, A.L.; Wang, D.; Foppen, E.; Jansen, R.; Boudzovitch-Surovtseva, O.; de Vries, J.; Fliers, E.; Kalsbeek, A. Feeding during the resting phase causes profound changes in physiology and desynchronization between liver and muscle rhythms of rats. Eur. J. Neurosci. 2016, 44, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- Brewer, M.; Lange, D.; Baler, R.; Anzulovich, A. SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. J. Biol. Rhythm. 2005, 20, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Ukena, K.; Iwakoshi-Ukena, E.; Taniuchi, S.; Bessho, Y.; Maejima, S.; Masuda, K.; Shikano, K.; Kondo, K.; Furumitsu, M.; Tachibana, T. Identification of a cDNA encoding a novel small secretory protein, neurosecretory protein GL, in the chicken hypothalamic infundibulum. Biochem. Biophys. Res. Commun. 2014, 446, 298–303. [Google Scholar] [CrossRef]
- Iwakoshi-Ukena, E.; Shikano, K.; Kondo, K.; Taniuchi, S.; Furumitsu, M.; Ochi, Y.; Sasaki, T.; Okamoto, S.; Bentley, G.E.; Kriegsfeld, L.J.; et al. Neurosecretory protein GL stimulates food intake, de novo lipogenesis, and onset of obesity. eLife 2017, 6, e28527. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, D.; Shikano, K.; Saito, T.; Iwakoshi-Ukena, E.; Furumitsu, M.; Ochi, Y.; Sato, M.; Bentley, G.E.; Kriegsfeld, L.J.; Ukena, K. Neurosecretory protein GL, a hypothalamic small secretory protein, participates in energy homeostasis in male mice. Endocrinology 2017, 158, 1120–1129. [Google Scholar] [CrossRef]
- Shikano, K.; Bessho, Y.; Kato, M.; Iwakoshi-Ukena, E.; Taniuchi, S.; Furumitsu, M.; Tachibana, T.; Bentley, G.E.; Kriegsfeld, L.J.; Ukena, K. Localization and function of neurosecretory protein GM, a novel small secretory protein, in the chicken hypothalamus. Sci. Rep. 2018, 8, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikano, K.; Iwakoshi-Ukena, E.; Saito, T.; Narimatsu, Y.; Kadota, A.; Furumitsu, M.; Bentley, G.E.; Kriegsfeld, L.J.; Ukena, K. Neurosecretory protein GL induces fat accumulation in mice. J. Endocrinol. 2020, 244, 1–12. [Google Scholar] [CrossRef]
- Shikano, K.; Kato, M.; Iwakoshi-Ukena, E.; Furumitsu, M.; Matsuura, D.; Masuda, K.; Tachibana, T.; Bentley, G.E.; Kriegsfeld, L.J.; Ukena, K. Effects of chronic intracerebroventricular infusion of neurosecretory protein GL on body mass and food and water intake in chicks. Gen. Comp. Endocrinol. 2018, 256, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Shikano, K.; Iwakoshi-Ukena, E.; Kato, M.; Furumitsu, M.; Bentley, G.E.; Kriegsfeld, L.J.; Ukena, K. Neurosecretory protein GL induces fat accumulation in chicks. Front. Endocrinol. 2019, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Arendt, J. Shift work: Coping with the biological clock. Occup. Med. 2010, 60, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Wulff, K.; Gatti, S.; Wettstein, J.G.; Foster, R.G. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010, 11, 589–599. [Google Scholar] [CrossRef]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef] [Green Version]
- Zvonic, S.; Ptitsyn, A.A.; Conrad, S.A.; Scott, L.K.; Floyd, Z.E.; Kilroy, G.; Wu, X.; Goh, B.C.; Mynatt, R.L.; Gimble, J.M. Characterization of peripheral circadian clocks in adipose tissues. Diabetes 2006, 55, 962–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, T.M.; Breininger, J.F.; Baskin, D.G.; Schwartz, M.W. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat. Neurosci. 1998, 1, 271–272. [Google Scholar] [CrossRef] [PubMed]
- Minokoshi, Y.; Alquier, T.; Furukawa, N.; Kim, Y.B.; Lee, A.; Xue, B.; Mu, J.; Foufelle, F.; Ferré, P.; Birnbaum, M.J.; et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 2004, 428, 569–574. [Google Scholar] [CrossRef]
- Um, J.H.; Pendergast, J.S.; Springer, D.A.; Foretz, M.; Viollet, B.; Brown, A.; Kim, M.K.; Yamazaki, S.; Chung, J.H. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS ONE 2011, 6, e18450. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Opperhuizen, A.L.; Reznick, J.; Turner, N.; Su, Y.; Cooney, G.J.; Kalsbeek, A. Effects of feeding time on daily rhythms of neuropeptide and clock gene expression in the rat hypothalamus. Brain Res. 2017, 1671, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Stephan, F.K. The other circadian system: Food as a Zeitgeber. J. Biol. Rhythm. 2002, 17, 284–292. [Google Scholar] [CrossRef]
- Antle, M.C.; Silver, R. Neural basis of timing and anticipatory behaviors. Eur. J. Neurosci. 2009, 30, 1643–1649. [Google Scholar] [CrossRef] [Green Version]
- Tahara, Y.; Hirao, A.; Moriya, T.; Kudo, T.; Shibata, S. Effects of medial hypothalamic lesions on feeding-induced entrainment of locomotor activity and liver Per2 expression in Per2::luc mice. J. Biol. Rhythm. 2010, 25, 9–18. [Google Scholar] [CrossRef]
- Davidson, A.J. Lesion studies targeting food-anticipatory activity. Eur. J. Neurosci. 2009, 30, 1658–1664. [Google Scholar] [CrossRef]
- Yamamuro, D.; Takahashi, M.; Nagashima, S.; Wakabayashi, T.; Yamazaki, H.; Takei, A.; Takei, S.; Sakai, K.; Ebihara, K.; Iwasaki, Y.; et al. Peripheral circadian rhythms in the liver and white adipose tissue of mice are attenuated by constant light and restored by time-restricted feeding. PLoS ONE 2020, 15, e0234439. [Google Scholar] [CrossRef] [PubMed]
- Karbowska, J.; Kochan, Z.; Swierczynski, J. Increase of lipogenic enzyme mRNA levels in rat white adipose tissue after multiple cycles of starvation-refeeding. Metabolism 2001, 50, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 6th ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Boston, MA, USA, 2007. [Google Scholar]
- Hughes, M.E.; Hogenesch, J.B.; Kornacker, K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythm. 2010, 25, 372–380. [Google Scholar] [CrossRef]
| Tissue | MBH | Liver | WAT | BAT | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | ALF | RF | ALF | RF | ALF | RF | ALF | RF |
| Bmal1 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Per2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Cry1 | 0.085 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Nr1d1 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Npgl | 0.003 | 0.020 | ||||||
| Npgm | <0.001 | 0.021 | ||||||
| Npy | <0.001 | 0.001 | ||||||
| Pomc | 0.187 | 0.166 | ||||||
| Mch | 0.045 | 0.483 | ||||||
| Orx | 0.063 | 0.083 | ||||||
| Acc | <0.001 | <0.001 | 0.004 | 0.899 | 0.299 | 0.004 | ||
| Fasn | 0.010 | <0.001 | <0.001 | 0.080 | 0.268 | 0.001 | ||
| Scd1 | <0.001 | <0.001 | 0.049 | 0.657 | 0.072 | 0.208 | ||
| Atgl | <0.001 | <0.001 | 0.089 | 0.280 | 0.125 | <0.001 | ||
| Cpt1a | <0.001 | <0.001 | <0.001 | 0.229 | 0.434 | <0.001 | ||
| Hsl | <0.001 | <0.001 | 0.004 | 0.002 | 0.043 | 0.058 | ||
| Tissue | Gene | p-Value | Amplitude | Acrophase | |||
|---|---|---|---|---|---|---|---|
| ALF | RF | ALF | RF | ALF | RF | ||
| MBH | Bmal1 | <0.001 | <0.001 | 0.157 | 0.117 | 0 | 12 |
| Per2 | <0.001 | <0.001 | 0.391 | 0.285 | 15 | 7.5 | |
| Cry1 | 0.021 | <0.001 | 0.095 | 0.093 | 16.5 | 10.5 | |
| Nr1d1 | <0.001 | <0.001 | 0.309 | 0.184 | 9 | 1.5 | |
| Npgl | 0.935 | 0.268 | 0.079 | 0.306 | 15 | 3 | |
| Npgm | 0.005 | 0.029 | 0.278 | 0.151 | 19.5 | 1.5 | |
| Npy | 1.000 | 0.338 | 0.046 | 0.041 | 21 | 7.5 | |
| Pomc | 1.000 | 0.002 | 0.106 | 0.166 | 12 | 15 | |
| Mch | 0.013 | 0.244 | 0.094 | 0.061 | 19.5 | 9 | |
| Orx | 0.241 | 0.098 | 0.237 | 0.167 | 18 | 12 | |
| Liver | Bmal1 | <0.001 | <0.001 | 0.520 | 0.732 | 22.5 | 15 |
| Per2 | <0.001 | <0.001 | 7.619 | 6.659 | 13.5 | 4.5 | |
| Cry1 | <0.001 | <0.001 | 0.460 | 0.313 | 19.5 | 10.5 | |
| Nr1d1 | <0.001 | <0.001 | 8.220 | 8.185 | 7.5 | 22.5 | |
| Acc | <0.001 | <0.001 | 0.797 | 0.852 | 13.5 | 4.5 | |
| Fasn | <0.001 | <0.001 | 0.859 | 1.212 | 15 | 7.5 | |
| Scd1 | 0.019 | <0.001 | 0.305 | 0.381 | 10.5 | 22.5 | |
| Atgl | <0.001 | <0.001 | 0.853 | 1.293 | 9 | 0 | |
| Cpt1a | <0.001 | <0.001 | 0.802 | 1.178 | 9 | 0 | |
| Hsl | <0.001 | <0.001 | 0.348 | 0.760 | 13.5 | 1.5 | |
| WAT | Bmal1 | <0.001 | <0.001 | 0.449 | 0.771 | 0 | 12 |
| Per2 | <0.001 | <0.001 | 2.582 | 1.594 | 13.5 | 6 | |
| Cry1 | <0.001 | <0.001 | 0.550 | 0.820 | 18 | 10.5 | |
| Nr1d1 | <0.001 | <0.001 | 1.727 | 1.765 | 7.5 | 22.5 | |
| Acc | 0.005 | 1.000 | 0.174 | 0.294 | 18 | 15 | |
| Fasn | <0.001 | 0.072 | 0.448 | 0.833 | 19.5 | 12 | |
| Scd1 | 0.052 | 0.946 | 0.128 | 0.202 | 18 | 1.5 | |
| Atgl | 0.014 | 0.011 | 0.217 | 0.202 | 13.5 | 1.5 | |
| Cpt1a | <0.001 | 0.668 | 0.299 | 0.049 | 7.5 | 18 | |
| Hsl | <0.001 | <0.001 | 0.306 | 0.495 | 12 | 0 | |
| BAT | Bmal1 | <0.001 | <0.001 | 0.409 | 0.582 | 0 | 13.5 |
| Per2 | <0.001 | 0.053 | 3.981 | 0.713 | 15 | 6 | |
| Cry1 | <0.001 | <0.001 | 0.374 | 0.648 | 18 | 13.5 | |
| Nr1d1 | <0.001 | <0.001 | 2.477 | 1.037 | 7.5 | 21 | |
| Acc | 1.000 | <0.001 | 0.083 | 0.910 | 6 | 12 | |
| Fasn | 1.000 | <0.001 | 0.122 | 3.861 | 21 | 12 | |
| Scd1 | 0.209 | 0.485 | 0.066 | 1.205 | 18 | 15 | |
| Atgl | 0.016 | 1.000 | 0.094 | 0.049 | 10.5 | 0 | |
| Cpt1a | 0.017 | <0.001 | 0.105 | 0.234 | 9 | 3 | |
| Hsl | 0.008 | 0.570 | 0.136 | 0.087 | 12 | 19.5 | |
| Tissue | Gene | Feeding | Time | Interaction | |||
|---|---|---|---|---|---|---|---|
| MBH | Bmal1 | F(1,70) = 31.85 | <0.001 | F(7,70) = 1.17 | 0.333 | F(7,70) = 10.62 | <0.001 |
| Per2 | F(1,70) = 0.08 | 0.779 | F(7,70) = 13.20 | <0.001 | F(7,70) = 12.18 | <0.001 | |
| Cry1 | F(1,70) = 0.56 | 0.473 | F(7,70) = 6.54 | <0.001 | F(7,70) = 8.67 | <0.001 | |
| Nr1d1 | F(1,70) = 1.20 | 0.299 | F(7,70) = 8.42 | <0.001 | F(7,70) = 10.52 | <0.001 | |
| Npgl | F(1,63) = 203.32 | <0.001 | F(7,63) = 2.59 | 0.021 | F(7,63) = 2.56 | 0.022 | |
| Npgm | F(1,56) = 9.68 | 0.014 | F(7,56) = 5.45 | <0.001 | F(7,56) = 2.71 | 0.017 | |
| Npy | F(1,63) = 0.83 | 0.386 | F(7,63) = 5.91 | <0.001 | F(7,63) = 2.98 | 0.009 | |
| Pomc | F(1,35) = 0.18 | 0.691 | F(7,35) = 1.55 | 0.185 | F(7,35) = 2.04 | 0.077 | |
| Mch | F(1,63) = 0.88 | 0.372 | F(7,63) = 0.93 | 0.488 | F(7,63) = 2.72 | 0.016 | |
| Orx | F(1,63) = 1.94 | 0.197 | F(7,63) = 4.33 | <0.001 | F(7,63) = 2.21 | 0.045 | |
| Liver | Bmal1 | F(1,70) = 96.04 | <0.001 | F(7,70) = 37.49 | <0.001 | F(7,70) = 120.42 | <0.001 |
| Per2 | F(1,70) = 3.58 | 0.088 | F(7,70) = 19.92 | <0.001 | F(7,70) = 110.12 | <0.001 | |
| Cry1 | F(1,70) = 0.31 | 0.589 | F(7,70) = 17.13 | <0.001 | F(7,70) = 45.10 | <0.001 | |
| Nr1d1 | F(1,70) = 4.99 | 0.049 | F(7,70) = 42.84 | <0.001 | F(7,70) = 142.87 | <0.001 | |
| Acc | F(1,70) = 1.40 | 0.263 | F(7,70) = 9.79 | <0.001 | F(7,70) = 24.55 | <0.001 | |
| Fasn | F(1,70) = 3.82 | 0.079 | F(7,70) = 9.34 | <0.001 | F(7,70) = 14.25 | <0.001 | |
| Scd1 | F(1,70) = 2.46 | 0.148 | F(7,70) = 2.98 | 0.009 | F(7,70) = 13.96 | <0.001 | |
| Atgl | F(1,70) = 10.18 | 0.010 | F(7,70) = 27.59 | <0.001 | F(7,70) = 92.00 | <0.001 | |
| Cpt1a | F(1,70) = 1.49 | 0.251 | F(7,70) = 26.36 | <0.001 | F(7,70) = 53.11 | <0.001 | |
| Hsl | F(1,70) = 2.86 | 0.122 | F(7,70) = 7.95 | <0.001 | F(7,70) = 33.51 | <0.001 | |
| WAT | Bmal1 | F(1,70) = 209.59 | <0.001 | F(7,70) = 22.07 | <0.001 | F(7,70) = 117.58 | <0.001 |
| Per2 | F(1,70) = 19.61 | 0.001 | F(7,70) = 15.23 | <0.001 | F(7,70) = 49.22 | <0.001 | |
| Cry1 | F(1,70) = 0.12 | 0.734 | F(7,70) = 22.43 | <0.001 | F(7,70) = 39.12 | <0.001 | |
| Nr1d1 | F(1,70) = 0.55 | 0.476 | F(7,70) = 6.28 | <0.001 | F(7,70) = 47.65 | <0.001 | |
| Acc | F(1,70) = 389.30 | <0.001 | F(7,70) = 0.59 | 0.761 | F(7,70) = 0.38 | 0.912 | |
| Fasn | F(1,70) = 215.27 | <0.001 | F(7,70) = 1.98 | 0.070 | F(7,70) = 2.17 | 0.048 | |
| Scd1 | F(1,63) = 923.63 | <0.001 | F(7,63) = 1.73 | 0.119 | F(7,63) = 2.04 | 0.063 | |
| Atgl | F(1,70) = 41.25 | <0.001 | F(7,70) = 0.66 | 0.705 | F(7,70) = 2.23 | 0.041 | |
| Cpt1a | F(1,70) = 22.35 | <0.001 | F(7,70) = 2.52 | 0.023 | F(7,70) = 5.59 | <0.001 | |
| Hsl | F(1,70) = 0.37 | 0.556 | F(7,70) = 0.65 | 0.710 | F(7,70) = 7.33 | <0.001 | |
| BAT | Bmal1 | F(1,70) = 58.84 | <0.001 | F(7,70) = 8.29 | <0.001 | F(7,70) = 87.98 | <0.001 |
| Per2 | F(1,63) = 20.23 | 0.001 | F(7,63) = 49.13 | <0.001 | F(7,63) = 56.73 | <0.001 | |
| Cry1 | F(1,63) = 23.80 | <0.001 | F(7,63) = 22.92 | <0.001 | F(7,63) = 25.43 | <0.001 | |
| Nr1d1 | F(1,70) = 1.22 | 0.295 | F(7,70) = 10.25 | <0.001 | F(7,70) = 49.35 | <0.001 | |
| Acc | F(1,70) = 1549.45 | <0.001 | F(7,70) = 3.20 | 0.005 | F(7,70) = 3.05 | 0.007 | |
| Fasn | F(1,70) = 399.79 | <0.001 | F(7,70) = 4.25 | <0.001 | F(7,70) = 4.13 | <0.001 | |
| Scd1 | F(1,70) = 509.92 | <0.001 | F(7,70) = 1.38 | 0.226 | F(7,70) = 1.36 | 0.235 | |
| Atgl | F(1,70) = 52.20 | <0.001 | F(7,70) = 3.50 | 0.003 | F(7,70) = 2.21 | 0.044 | |
| Cpt1a | F(1,70) = 4.10 | 0.070 | F(7,70) = 2.21 | 0.044 | F(7,70) = 2.55 | 0.021 | |
| Hsl | F(1,70) = 11.44 | 0.007 | F(7,70) = 2.96 | 0.009 | F(7,70) = 2.65 | 0.017 | |
| Gene | Sense Primer (5’ to 3’) | Antisense Primer (5’ to 3’) |
|---|---|---|
| Bmal1 | ACATAGGACACCTCGCAGAA | AACCATCGACTTCGTAGCGT |
| Per2 | TCTGACATGGCTTCTGTTCG | TGTACAGTGTGGGGGTGCTA |
| Cry1 | GGGACAGCCAGCTGATGTAT | CATCTCGTTCCTTCCCAAAA |
| Nr1d1 | AGCCACCCCAAGACCTTACT | CGGTCATTCAAACTGGACCT |
| Npgl | GGAACCATGGCTTAGGAAGG | TCTAAGGAGCTGAGAATATGCA |
| Npgm | CTCTCTGACGCTGATAGACC | AGATACTGTAATGCCCAGGA |
| Npy | TATCTCTGCTCGTGTGTTTG | GATTGATGTAGTGTCGCAGA |
| Pomc | AGCTGCCTTTCCGCGACA | ATCTATGGAGGTCTGAAGCA |
| Mch | GGAAGGAGAGATTTTGACATGCTC | TTCTTCTGTAAGGATGTTGCGGAC |
| Orx | GCCTCCTTCAGGCCAACGGTAA | GGGGTGCTAAAGCGGTGGTAGT |
| Acc | TCCGCACTGACTGTAACCACAT | TGCTCCGCACAGATTCTTCA |
| Fasn | AGGGGTCGACCTGGTCCTCA | GCCATGCCCAGAGGGTGGTT |
| Scd1 | CTGTACGGGATCATACTGGTTC | GCCGTGCCTTGTAAGTTCTG |
| Atgl | AACACCAGCATCCAGTTCAA | GGTTCAGTAGGCCATTCCTC |
| Cpt1a | CCTGGGCATGATTGCAAAG | GGACGCCACTCACGATGTT |
| Hsl | GCTGGGCTGTCAAGCACTGT | GTAACTGGGTAGGCTGCCAT |
| Actb | GGCACCACACCTTCTACAAT | AGGTCTCAAACATGATCTGG |
| Rps18 | CCTGAGAAGTTCCAGCACAT | TTCTCCAGCCCTCTTGGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadota, A.; Iwakoshi-Ukena, E.; Fukumura, K.; Shikano, K.; Narimatsu, Y.; Furumitsu, M.; Ukena, K. Effects of Irregular Feeding on the Daily Fluctuations in mRNA Expression of the Neurosecretory Protein GL and Neurosecretory Protein GM Genes in the Mouse Hypothalamus. Int. J. Mol. Sci. 2021, 22, 2109. https://doi.org/10.3390/ijms22042109
Kadota A, Iwakoshi-Ukena E, Fukumura K, Shikano K, Narimatsu Y, Furumitsu M, Ukena K. Effects of Irregular Feeding on the Daily Fluctuations in mRNA Expression of the Neurosecretory Protein GL and Neurosecretory Protein GM Genes in the Mouse Hypothalamus. International Journal of Molecular Sciences. 2021; 22(4):2109. https://doi.org/10.3390/ijms22042109
Chicago/Turabian StyleKadota, Atsuki, Eiko Iwakoshi-Ukena, Keisuke Fukumura, Kenshiro Shikano, Yuki Narimatsu, Megumi Furumitsu, and Kazuyoshi Ukena. 2021. "Effects of Irregular Feeding on the Daily Fluctuations in mRNA Expression of the Neurosecretory Protein GL and Neurosecretory Protein GM Genes in the Mouse Hypothalamus" International Journal of Molecular Sciences 22, no. 4: 2109. https://doi.org/10.3390/ijms22042109






