Water Extract of Mungbean (Vigna radiata L.) Inhibits Protein Tyrosine Phosphatase-1B in Insulin-Resistant HepG2 Cells
Abstract
:1. Introduction
2. Results
2.1. Total Phenolic Content and Flavonoid Identification of MWE
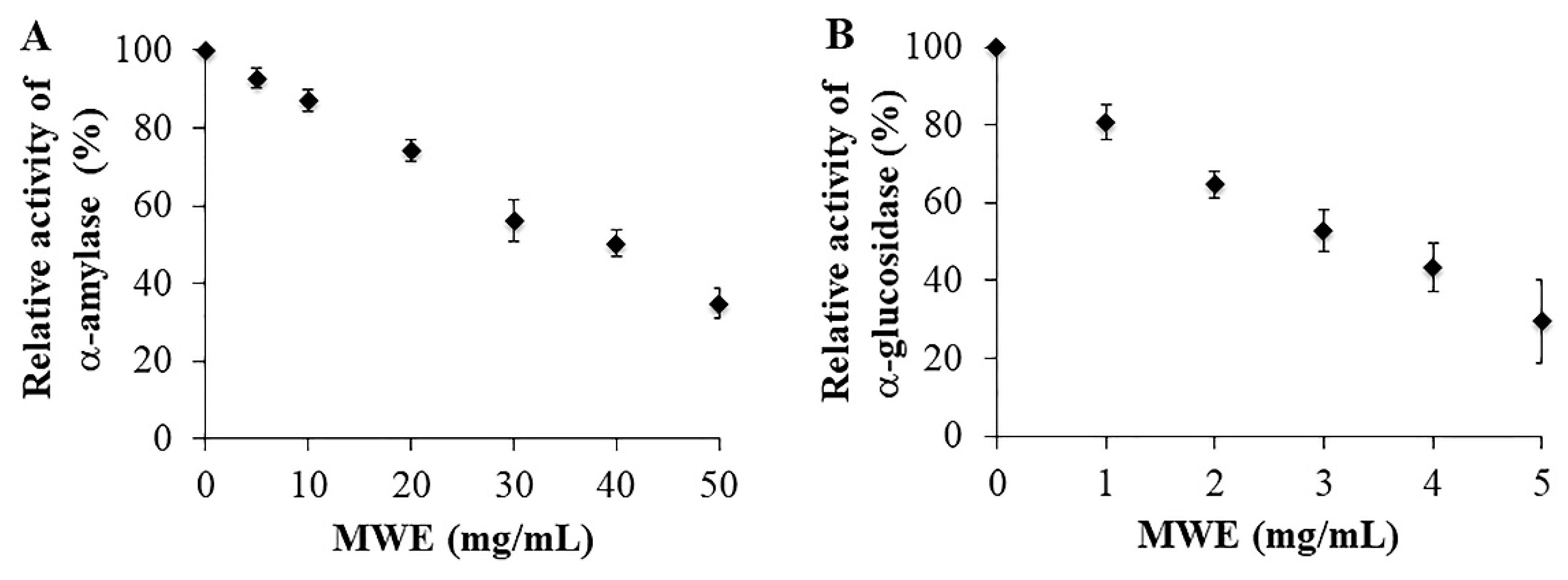
2.2. Inhibitory Effects of MWE against A-Amylase and A-Glucosidase
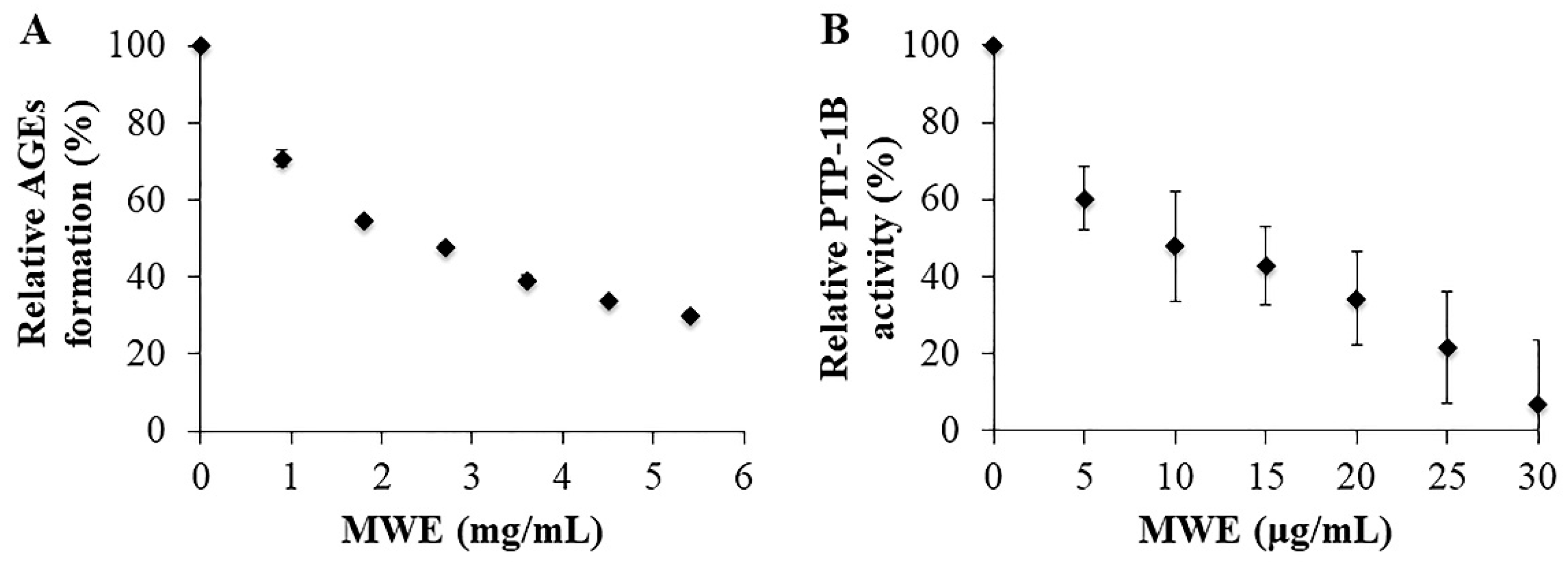
2.3. Inhibitory Effects of MWE against AGEs Formation
2.4. Inhibitory Effects of MWE against PTP-1B
2.5. The Effect of MWE on Cytotoxicity at 24 H Of Treatment in Hepg2 Cells
2.6. The Effect of MWE on Cellular Glucose Uptake of Insulin-Resistant Hepg2 Cells
2.7. The Effect of MWE on the Expression of Genes Related to Glucose Metabolism in Insulin-Resistant Hepg2 Cells
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Preparation of Mungbean Water Extract (MWE)
4.3. Determination of Total Phenolic Content
4.4. Separation and Identification of Flavonoids Using NMR
4.5. Standardization of Major Flavonoids in MWE
4.6. Assay of A-Amylase Inhibition
4.7. Assay of α-Glucosidase Inhibition
4.8. Inhibition of Advanced Glycation End Products (Ages) Formation Assay
4.9. Protein Tyrosine Phosphatase 1B (PTP-1B) Inhibition Assay
4.10. Cell culture and Cytotoxicity Assay
4.11. Cellular Glucose Uptake Assay
4.12. Gene Expression of Insulin-Resistant Hepg2 Cells
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| ACE | angiotensin I-converting enzyme |
| AG | aminoguanidine |
| AGEs | advanced glycation end products |
| ANOVA | analysis of variance |
| AU | arbitrary unit |
| BSA | bovine serum albumin |
| DAD | diode array detector |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO-d6 | deuterated dimethylsulfoxide |
| DNS | dinitrosalicylic acid |
| DW | dry weight |
| EGCG | (-)-epigallocatechin-3-gallate |
| FOXO1 | forkhead box O1 |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| GAE | gallic acid equivalent |
| GSK-3β | glycogene synthase kinase-3β |
| HPLC | high-performance liquid chromatography |
| IR | insulin-resistant HepG2 cells |
| MWE | mungbean water extract |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetra-zolium bromide |
| NaN3 | sodium azide |
| 2-NBDG | 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose |
| NMR | nuclear magnetic resonance |
| PCR | polymerase chain reaction |
| PEPCK | phosphoenolpyruvate carboxykinase |
| PI-3K | phosphatidylinosital 3-kinase |
| PKB/Akt | protein kinase B |
| pNP | p-nitrophenyl |
| pNPG | para-nirophenyl-α-d-glucopyranoside |
| pNPP | p-nitrophenyl phosphate |
| PTP-1B | protein tyrosine phosphatase-1B |
| UV | ultraviolet |
References
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; da Rocha Fernandes, J.D.; Ohlrogge, A.W.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pr. 2018, 138, 271–281. [Google Scholar] [CrossRef]
- Dong, X.C.; Copps, K.D.; Guo, S.; Li, Y.; Kollipara, R.; DePinho, R.A.; White, M.F. Inactivation of Hepatic Foxo1 by Insulin Signaling Is Required for Adaptive Nutrient Homeostasis and Endocrine Growth Regulation. Cell Metab. 2008, 8, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pr 2019, 157, 107843. [Google Scholar] [CrossRef] [Green Version]
- Manukumar, H.M.; Kumar, J.S.; Chandrasekhar, B.; Raghava, S.; Umesha, S. Evidences for diabetes and insulin mimetic activity of medicinal plants: Present status and future prospects. Crit. Rev. Food Sci. 2017, 57, 2712–2729. [Google Scholar] [CrossRef]
- Fei, Q.; Gao, Y.; Zhang, X.; Sun, Y.; Hu, B.; Zhou, L.; Jabbar, S.; Zeng, X. Effects of Oolong Tea Polyphenols, EGCG, and EGCG3″Me on Pancreatic α-Amylase Activity in Vitro. J. Agric. Food Chem. 2014, 62, 9507–9514. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Cheng, X.Z.; Wang, L.X.; Wang, S.H.; Ren, G.X. A Determination of Potential alpha-Glucosidase Inhibitors from Azuki Beans (Vigna angularis). Int. J. Mol. Sci. 2011, 12, 6445–6451. [Google Scholar] [CrossRef] [Green Version]
- Derosa, G.; Maffioli, P. α-Glucosidase inhibitors and their use in clinical practice. Arch Med Sci 2012, 8, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Farsi, E.; Shafaei, A.; Hor, S.Y.; Khadeer Ahamed, M.B.; Yam, M.F.; Attitalla, I.H.; Asmawi, M.Z.; Ismail, Z. Correlation between enzymes inhibitory effects and antioxidant activities of standardized fractions of methanolic extract obtained from Ficus deltoidea leaves. Afr. J. Biotechnol. 2011, 10, 15184–15194. [Google Scholar] [CrossRef] [Green Version]
- Adeva-Andany, M.M.; Pérez-Felpete, N.; Fernández-Fernández, C.; Donapetry-García, C.; Pazos-García, C. Liver glucose metabolism in humans. Biosci. Rep. 2016, 36, e00416. [Google Scholar] [CrossRef] [Green Version]
- Michael, M.D.; Kulkarni, R.N.; Postic, C.; Previs, S.F.; Shulman, G.I.; Magnuson, M.A.; Kahn, C.R. Loss of Insulin Signaling in Hepatocytes Leads to Severe Insulin Resistance and Progressive Hepatic Dysfunction. Mol. Cell 2000, 6, 87–97. [Google Scholar] [CrossRef]
- Gum, R.J.; Gaede, L.L.; Koterski, S.L.; Heindel, M.; Clampit, J.E.; Zinker, B.A.; Trevillyan, J.M.; Ulrich, R.G.; Jirousek, M.R.; Rondinone, C.M. Reduction of protein tyrosine phosphatase 1B increases insulin-dependent signaling in ob/ob mice. Diabetes 2003, 52, 21–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, S.; Fantus, I.G. Inhibition of the protein tyrosine phosphatase PTP1B: Potential therapy for obesity, insulin resistance and type-2 diabetes mellitus. Best Pract. Res. Clin. Endocrinol. Metab. 2007, 21, 621–640. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Guo, S.; Copps, K.; Dong, X.; Kollipara, R.; Rodgers, J.T.; Depinho, R.A.; Puigserver, P.; White, M.F. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat. Med. 2009, 15, 1307. [Google Scholar] [CrossRef] [Green Version]
- Goh, S.-Y.; Cooper, M.E. The Role of Advanced Glycation End Products in Progression and Complications of Diabetes. J. Clin. Endocrinol. Metab. 2008, 93, 1143–1152. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Zheng, Z.; Cheng, K.W.; Shan, F.; Ren, G.X.; Chen, F.; Wang, M. Inhibitory effect of mung bean extract and its constituents vitexin and isovitexin on the formation of advanced glycation endproducts. Food Chem. 2008, 106, 475–481. [Google Scholar] [CrossRef]
- Li, G.H.; Wan, J.Z.; Le, G.W.; Shi, Y.H. Novel angiotensin I-converting enzyme inhibitory peptides isolated from Alcalase hydrolysate of mung bean protein. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2006, 12, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.; Choi, S.; Ha, T.J.; Choi, M.; Wi, H.R.; Lee, B.W.; Lee, M. Effects of Mung Bean (Vigna radiata L.) Ethanol Extracts Decrease Proinflammatory Cytokine-Induced Lipogenesis in the KK-Ay Diabese Mouse Model. J. Med. Food 2015, 18, 841–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, H.; Inaba, Y.; Kimura, K.; Asahara, S.; Kido, Y.; Matsumoto, M.; Motoyama, T.; Tachibana, N.; Kaneko, S.; Kohno, M.; et al. Dietary mung bean protein reduces hepatic steatosis, fibrosis, and inflammation in male mice with diet-induced, nonalcoholic fatty liver disease. J. Nutr. 2016, 147, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Sae-tan, S.; Kumrungsee, T.; Yanaka, N. Mungbean seed coat water extract inhibits inflammation in LPS-induced acute liver injury mice and LPS-stimulated RAW 246.7 macrophages via the inhibition of TAK1/IκBα/NF-κB. J. Food Sci. Technol. 2020, 57, 2659–2668. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.H.; Kang, M.J.; Choe, E.O.; Shin, M.; Kim, J.I. Mung bean coat ameliorates hyperglycemia and the antioxidant status in type 2 diabetic db/db mice. Food Sci. Biotechnol. 2014, 23, 247–252. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, F.; Wang, M.; Wang, J.; Ren, G. Antidiabetic activity of Mung bean extracts in diabetic KK-Ay mice. J. Agric. Food Chem. 2008, 56, 8869–8873. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-P.; Li, Z.-Y.; Jia, J.-P.; Qin, X.-M. Chemical comparison of coat and kernel of mung bean by nuclear magnetic resonance-based metabolic fingerprinting approach. Spectrosc. Lett. 2016, 49, 217–224. [Google Scholar] [CrossRef]
- Luo, J.Q.; Cai, W.X.; Wu, T.; Xu, B.J. Phytochemical distribution in hull and cotyledon of adzuki bean (Vigna angularis L.) and mung bean (Vigna radiate L.), and their contribution to antioxidant, anti-inflammatory and anti-diabetic activities. Food Chem. 2016, 201, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, T.; Yun, Y.S.; Kunugi, A. Five novel flavonoids from Wasabia japonica. Tetrahedron 2005, 61, 7037–7044. [Google Scholar] [CrossRef]
- Huyer, G.; Liu, S.; Kelly, J.; Moffat, J.; Payette, P.; Kennedy, B.; Tsaprailis, G.; Gresser, M.J.; Ramachandran, C. Mechanism of Inhibition of Protein-tyrosine Phosphatases by Vanadate and Pervanadate*. J. Biol. Chem. 1997, 272, 843–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genet, S.; Kale, R.K.; Baquer, N.Z. Alterations in antioxidant enzymes and oxidative damage in experimental diabetic rat tissues: Effect of vanadate and fenugreek (Trigonella foenum graecum). Mol. Cell. Biochem. 2002, 236, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, S.; Xie, J.; Chen, Y.; Dong, R.; Zhang, X.; Liu, S.; Xie, J.; Hu, X.; Yu, Q. Antioxidant, α-amylase and α-glucosidase inhibitory activities of bound polyphenols extracted from mung bean skin dietary fiber. LWT 2020, 132, 109943. [Google Scholar] [CrossRef]
- Freedman, B.I.; Wuerth, J.-P.; Cartwright, K.; Bain, R.P.; Dippe, S.; Hershon, K.; Mooradian, A.D.; Spinowitz, B.S. Design and Baseline Characteristics for the Aminoguanidine Clinical Trial in Overt Type 2 Diabetic Nephropathy (ACTION II). Control. Clin. Trials 1999, 20, 493–510. [Google Scholar] [CrossRef]
- Thornalley, P.J. Use of aminoguanidine (Pimagedine) to prevent the formation of advanced glycation endproducts. Arch. Biochem. Biophys. 2003, 419, 31–40. [Google Scholar] [CrossRef]
- Bakke, J.; Haj, F.G. Protein-tyrosine phosphatase 1B substrates and metabolic regulation. Semin. Cell Dev. Biol. 2015, 37, 58–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haj, F.G.; Zabolotny, J.M.; Kim, Y.B.; Kahn, B.B.; Neel, B.G. Liver-specific protein-tyrosine phosphatase 1B (PTP1B) re-expression alters glucose Homeostasis of PTP1B-/- mice. J. Biol. Chem. 2005, 280, 15038–15046. [Google Scholar] [CrossRef] [Green Version]
- Mi, Y.; Zhang, W.; Tian, H.; Li, R.; Huang, S.; Li, X.; Qi, G.; Liu, X. EGCG evokes Nrf2 nuclear translocation and dampens PTP1B expression to ameliorate metabolic misalignment under insulin resistance condition. Food Funct. 2018, 9, 1510–1523. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Nurul Islam, M.; Yousof Ali, M.; Kim, E.J.; Kim, Y.M.; Jung, H.A. Effects of C-glycosylation on anti-diabetic, anti-Alzheimer’s disease and anti-inflammatory potential of apigenin. Food Chem. Toxicol. 2014, 64, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Herrera, I.; Martín, M.Á.; Goya, L.; Ramos, S. Cocoa flavonoids attenuate high glucose-induced insulin signalling blockade and modulate glucose uptake and production in human HepG2 cells. Food Chem. Toxicol. 2014, 64, 10–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Cao, D.; Yi, J.; Cao, J.; Jiang, W. Identification of the flavonoids in mungbean (Phaseolus radiatus L.) soup and their antioxidant activities. Food Chem. 2012, 135, 2942–2946. [Google Scholar] [CrossRef]
- Lekshmi, P.C.; Arimboor, R.; Indulekha, P.S.; Nirmala Menon, A. Turmeric (Curcuma longa L.) volatile oil inhibits key enzymes linked to type 2 diabetes. Int. J. Food Sci. Nutr. 2012, 63, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Wang, Y.; Shen, Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J. Biochem. Biophys. Methods 2005, 64, 207–215. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeting, O.; Chandarajoti, K.; Phongphisutthinan, A.; Hongsprabhas, P.; Sae-tan, S. Water Extract of Mungbean (Vigna radiata L.) Inhibits Protein Tyrosine Phosphatase-1B in Insulin-Resistant HepG2 Cells. Molecules 2021, 26, 1452. https://doi.org/10.3390/molecules26051452
Saeting O, Chandarajoti K, Phongphisutthinan A, Hongsprabhas P, Sae-tan S. Water Extract of Mungbean (Vigna radiata L.) Inhibits Protein Tyrosine Phosphatase-1B in Insulin-Resistant HepG2 Cells. Molecules. 2021; 26(5):1452. https://doi.org/10.3390/molecules26051452
Chicago/Turabian StyleSaeting, Orathai, Kasemsiri Chandarajoti, Angsuma Phongphisutthinan, Parichat Hongsprabhas, and Sudathip Sae-tan. 2021. "Water Extract of Mungbean (Vigna radiata L.) Inhibits Protein Tyrosine Phosphatase-1B in Insulin-Resistant HepG2 Cells" Molecules 26, no. 5: 1452. https://doi.org/10.3390/molecules26051452






