Phytogenic Water Additives Improve Broiler Growth Performance via Modulation of Intermediary Metabolism-Related Signaling Pathways
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animal Husbandry and Experimental Design
2.3. RNA Isolation and RT-qPCR
2.4. Western Blot Analysis
2.5. Statistical Analysis
3. Results
3.1. Hepatic Lipogenesis-Related Pathways
3.2. Lipolysis-Related Genes and Proteins in Adipose Tissue
3.3. Protein Synthesis-Related Pathway in Breast Muscle
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, G.; Hao, H.; Xie, S.; Wang, X.; Dai, M.; Huang, L.; Yuan, Z. Antibiotic alternatives: The substitution of antibiotics in animal husbandry? Front. Microbiol. 2014, 5, 217. [Google Scholar] [CrossRef] [Green Version]
- Gadde, U.; Kim, W.H.; Oh, S.T.; Lillehoj, H.S. Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: A review. Anim. Heal. Res. Rev. 2017, 18, 26–45. [Google Scholar] [CrossRef]
- Emami, N.K.; Calik, A.; White, M.B.; Young, M.; Dalloul, R.A. Necrotic Enteritis in Broiler Chickens: The Role of Tight Junctions and Mucosal Immune Responses in Alleviating the Effect of the Disease. Microorganisms 2019, 7, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emami, N.K.; Calik, A.; White, M.B.; Kimminau, E.A.; Dalloul, R.A. Effect of Probiotics and Multi-Component Feed Additives on Microbiota, Gut Barrier and Immune Responses in Broiler Chickens During Subclinical Necrotic Enteritis. Front. Veter-Sci. 2020, 7, 572142. [Google Scholar] [CrossRef] [PubMed]
- Windisch, W.; Schedle, K.; Plitzner, C.; Kroismayr, A. Use of phytogenic products as feed additives for swine and poultry1. J. Anim. Sci. 2008, 86, E140–E148. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.; Davoodi, H. Phytogenics as New Class of Feed Additive in Poultry Industry. J. Anim. Veter-Adv. 2010, 9, 2295–2304. [Google Scholar] [CrossRef]
- Emami, N.K.; Daneshmand, A.; Naeini, S.Z.; Graystone, E.N.; Broom, L.J. Effects of commercial organic acid blends on male broilers challenged with E. coli K88: Performance, microbiology, intestinal morphology, and immune respons. Poult. Sci. 2017, 96, 3254–3263. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, G.R.; Syed, B.; Haldar, S.; Pender, C. Phytogenic feed additives as an alternative to antibiotic growth promoters in broiler chickens. Front. Vet. Sci. 2015, 2, 21. [Google Scholar]
- Zhao, X.H.; He, X.; Yang, X.F.; Zhong, X.H. Effect of Portulaca oleracea extracts on growth performance and microbial populations in ceca of broilers. Poult. Sci. 2013, 92, 1343–1347. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Piao, X.S.; Zhang, Q.; Li, P.; Yi, J.Q.; Liu, J.D.; Li, Q.Y.; Wang, G.Q. The effects of Forsythia suspensa extract and berberine on growth performance, immunity, antioxidant activities, and intestinal microbiota in broilers under high stocking density. Poult. Sci. 2013, 92, 1981–1988. [Google Scholar] [CrossRef]
- Placha, I.; Takacova, J.; Ryzner, M.; Cobanova, K.; Laukova, A.; Strompfova, V.; Venglovska, K.; Faix, S. Effect of thyme essential oil and selenium on intestine integrity and antioxidant status of broilers. Br. Poult. Sci. 2014, 55, 105–114. [Google Scholar] [CrossRef]
- Ghazanfari, S.; Mohammadi, Z.; Moradi, M.A. Effects of Coriander Essential Oil on the Performance, Blood Characteristics, Intestinal Microbiota and Histological of Broilers. Braz. J. Poult. Sci. 2015, 17, 419–426. [Google Scholar] [CrossRef] [Green Version]
- Amad, A.A.; Männer, K.; Wendler, K.R.; Neumann, K.; Zentek, J. Effects of a phytogenic feed additive on growth performance and ileal nutrient digestibility in broiler chickens. Poult. Sci. 2011, 90, 2811–2816. [Google Scholar] [CrossRef]
- Flees, J.; Greene, E.; Ganguly, B.; Dridi, S. Phytogenic feed- and water-additives improve feed efficiency in broilers via modulation of (an)orexigenic hypothalamic neuropeptide expression. Neuropeptides 2020, 81, 102005. [Google Scholar] [CrossRef]
- Orlowski, S.; Flees, J.; Greene, E.S.; Ashley, D.; Lee, S.-O.; Yang, F.L.; Owens, C.M.; Kidd, M.; Anthony, N.; Dridi, S. Effects of phytogenic additives on meat quality traits in broiler chickens1. J. Anim. Sci. 2018, 96, 3757–3767. [Google Scholar] [CrossRef] [PubMed]
- Lassiter, K.; Greene, E.; Piekarski, A.; Faulkner, O.B.; Hargis, B.M.; Bottje, W.; Dridi, S. Orexin system is expressed in avian muscle cells and regulates mitochondrial dynamics. Am. J. Physiol. Integr. Comp. Physiol. 2015, 308, R173–R187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajaei-Sharifabadi, H.; Greene, E.; Piekarski, A.; Falcon, D.; Ellestad, L.; Donoghue, A.; Bottje, W.; Porter, T.; Liang, Y.; Dridi, S. Surface wetting strategy prevents acute heat exposure–induced alterations of hypothalamic stress–and metabolic-related genes in broiler chickens. J. Anim. Sci. 2017, 95, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.; Greene, E.; Ishola, P.; Huff, G.; Donoghue, A.; Bottje, W.; Dridi, S. Chronic Mild Cold Conditioning Modulates the Expression of Hypothalamic Neuropeptide and Intermediary Metabolic-Related Genes and Improves Growth Performances in Young Chicks. PLoS ONE 2015, 10, e0142319. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, K.; Gilley, A.; Piekarski, A.; Orlowski, S.; Greene, E.; Bottje, W.; Anthony, N.; Dridi, S.; Information, P.E.K.F.C. Differential expression of feeding-related hypothalamic neuropeptides in the first generation of quails divergently selected for low or high feed efficiency. Neuropeptides 2016, 58, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Flees, J.; Rajaei-Sharifabadi, H.; Greene, E.; Beer, L.; Hargis, B.M.; Ellestad, L.; Porter, T.; Donoghue, A.; Bottje, W.G.; Dridi, S. Effect of Morinda citrifolia (Noni)-Enriched Diet on Hepatic Heat Shock Protein and Lipid Metabolism-Related Genes in Heat Stressed Broiler Chickens. Front. Physiol. 2017, 8, 919. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hillgartner, F.B.; Salati, L.M.; Goodridge, A.G. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol. Rev. 1995, 75, 47–76. [Google Scholar] [CrossRef]
- Goodridge, A.G.; Garay, A.; Silpananta, P. Regulation of Lipogenesis and the Total Activities of Lipogenic Enzymes in a Primary Culture of Hepatocytes from Prenatal and Early Postnatal Chicks. J. Biol. Chem. 1974, 249, 1469–1475. [Google Scholar] [CrossRef]
- Leveille, G.A.; O’Hea, E.K.; Chakbabarty, K.; Chakrabarty, K. In Vivo Lipogenesis in the Domestic Chicken. Exp. Biol. Med. 1968, 128, 398–401. [Google Scholar] [CrossRef]
- Brownsey, R.; Zhande, R.; Boone, A. Isoforms of Acetyl-CoA Carboxylase: Structures, Regulatory Properties and Metabolic Functions. Biochem. Soc. Trans. 1997, 25, 1232–1238. [Google Scholar] [CrossRef]
- Tong, L. Acetyl-coenzyme A carboxylase: Crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. 2005, 62, 1784–1803. [Google Scholar] [CrossRef]
- Ha, J.; Daniel, S.; Broyles, S.S.; Kim, K.H. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J. Biol. Chem. 1994, 269, 22162–22168. [Google Scholar] [CrossRef]
- Jensen, K.K.; Tadin-Strapps, M.; Wang, S.P.; Hubert, J.; Kan, Y.; Ma, Y.; McLaren, D.G.; Previs, S.F.; Herath, K.B.; Mahsut, A.; et al. Dose-dependent effects of siRNA-mediated inhibition of SCAP on PCSK9, LDLR, and plasma lipids in mouse and rhesus monkey. J. Lipid Res. 2016, 57, 2150–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddineni, S.; Metzger, S.; Ocón, O.; Hendricks, G.; Ramachandran, R. Adiponectin Gene Is Expressed in Multiple Tissues in the Chicken: Food Deprivation Influences Adiponectin Messenger Ribonucleic Acid Expression. Endocrinology 2005, 146, 4250–4256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahmoorespur, M.; Ghazanfari, S.; Nobari, K. Evaluation of adiponectin gene expression in the abdominal adipose tissue of broiler chickens: Feed restriction, dietary energy, and protein influences adiponectin messenger ribonucleic acid expression. Poult. Sci. 2010, 89, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Gan, L.; Chen, D.; Sun, C. Adiponectin Impairs Chicken Preadipocytes Differentiation through p38 MAPK/ATF-2 and TOR/p70 S6 Kinase Pathways. PLoS ONE 2013, 8, e77716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonas, M.I.; Kurylowicz, A.; Bartoszewicz, Z.; Lisik, W.; Jonas, M.; Domienik-Karlowicz, J.; Puzianowska-Kuznicka, M. Adiponectin/resistin interplay in serum and in adipose tissue of obese and normal-weight individuals. Diabetol. Metab. Syndr. 2017, 9, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomer, X.; Pizarro-Delgado, J.; Barroso, E.; Vázquez-Carrera, M. Palmitic and Oleic Acid: The Yin and Yang of Fatty Acids in Type 2 Diabetes Mellitus. Trends Endocrinol. Metab. 2018, 29, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Piccinin, E.; Cariello, M.; De Santis, S.; Ducheix, S.; Sabbà, C.; Ntambi, J.M.; Moschetta, A. Role of Oleic Acid in the Gut-Liver Axis: From Diet to the Regulation of Its Synthesis via Stearoyl-CoA Desaturase 1 (SCD1). Nutrients 2019, 11, 2283. [Google Scholar] [CrossRef] [Green Version]
- Kraemer, F.B.; Shen, W.J. Hormone-sensitive lipase: Control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J. Lipid. Res. 2002, 43, 1585–1594. [Google Scholar] [CrossRef] [Green Version]
- Lucas, S.; Tavernier, G.; Tiraby, C.; Mairal, A.; Langin, D. Expression of human hormone-sensitive lipase in white adipose tissue of transgenic mice increases lipase activity but does not enhance in vitro lipolysis. J. Lipid. Res. 2003, 44, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Reid, B.N.; Ables, G.P.; Ottivanchik, O.A.; Schoiswohl, G.; Zechner, R.; Blaner, W.S.; Goldberg, I.J.; Schwabe, R.F.; Chua, S.C.; Huang, L.S. Hepatic Overexpression of Hormone-sensitive Lipase and Adipose Triglyceride Lipase Promotes Fatty Acid Oxidation Stimulates Direct Release of Free Fatty Acids, and Ameliorates Steatosis. J. Biol. Chem. 2008, 283, 13087–13099. [Google Scholar] [CrossRef] [Green Version]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell. Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef]
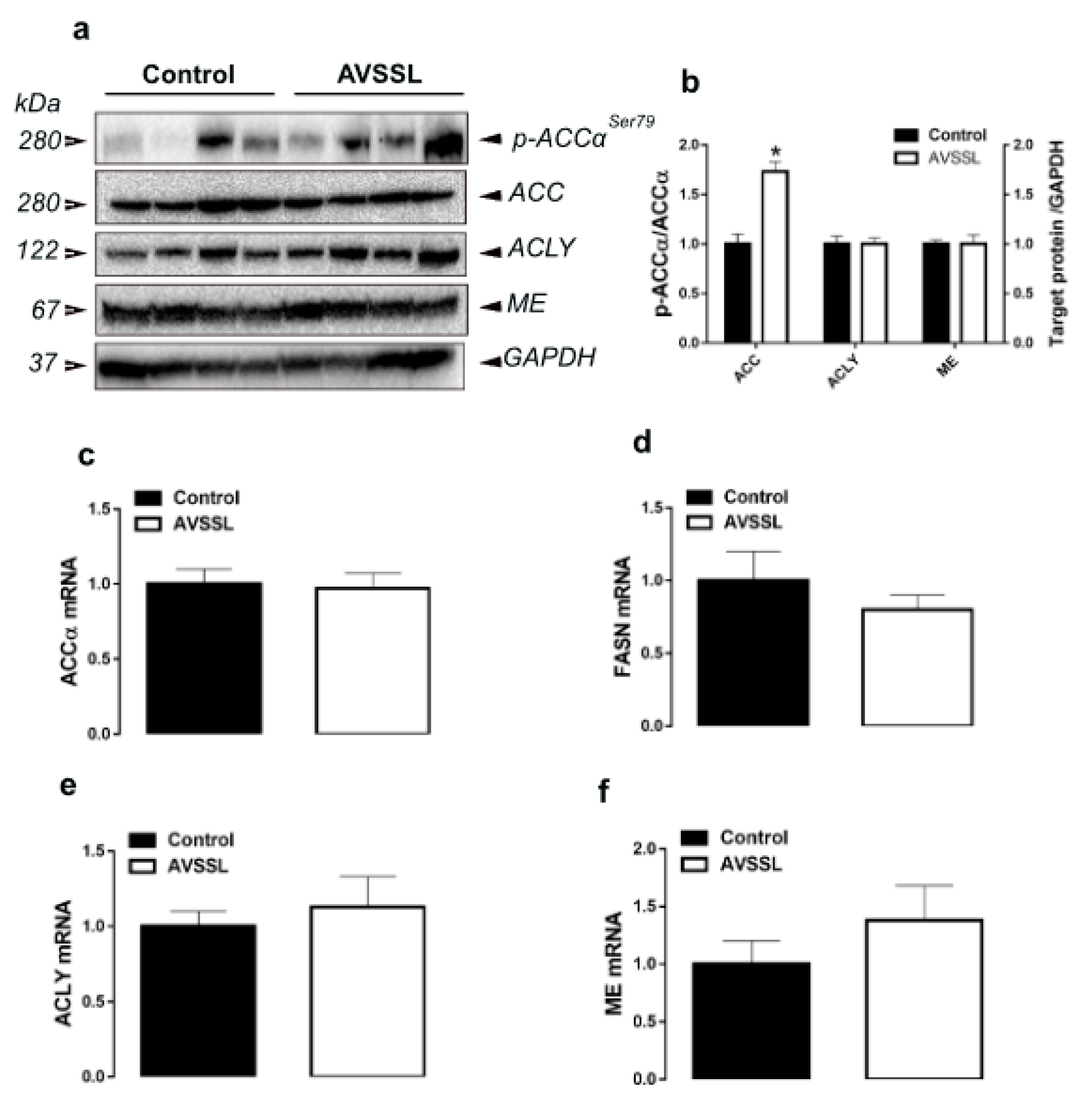
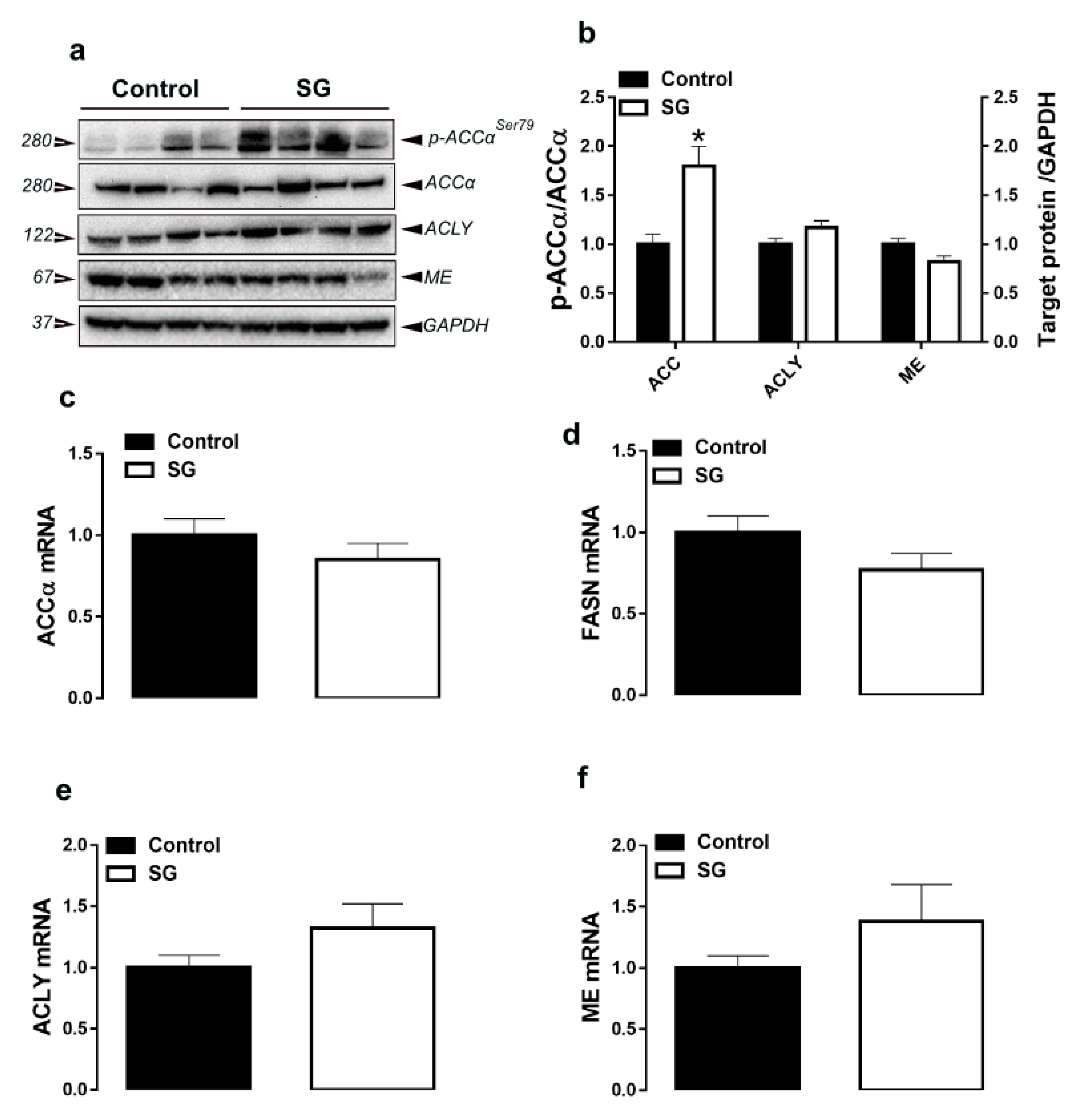
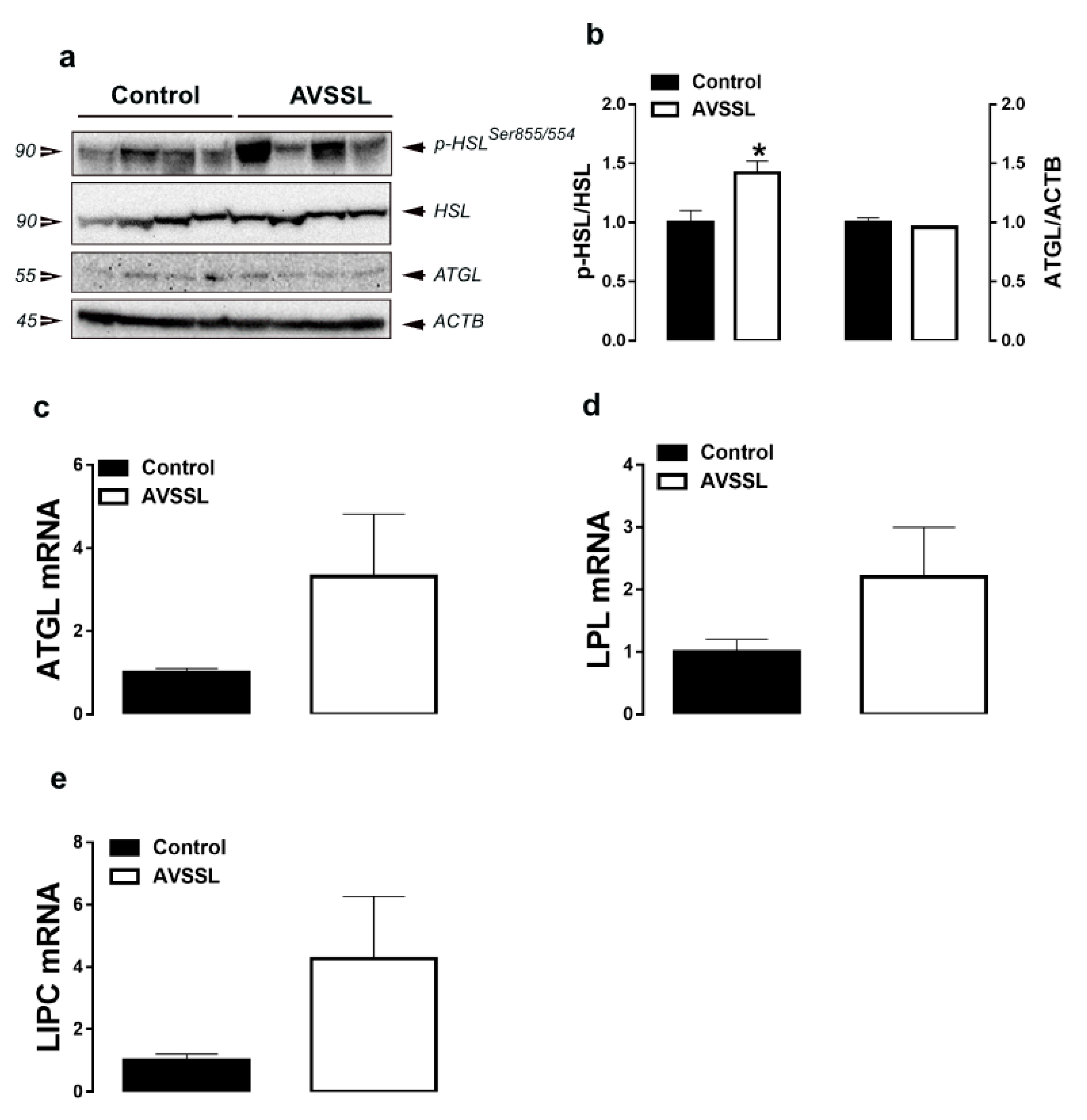
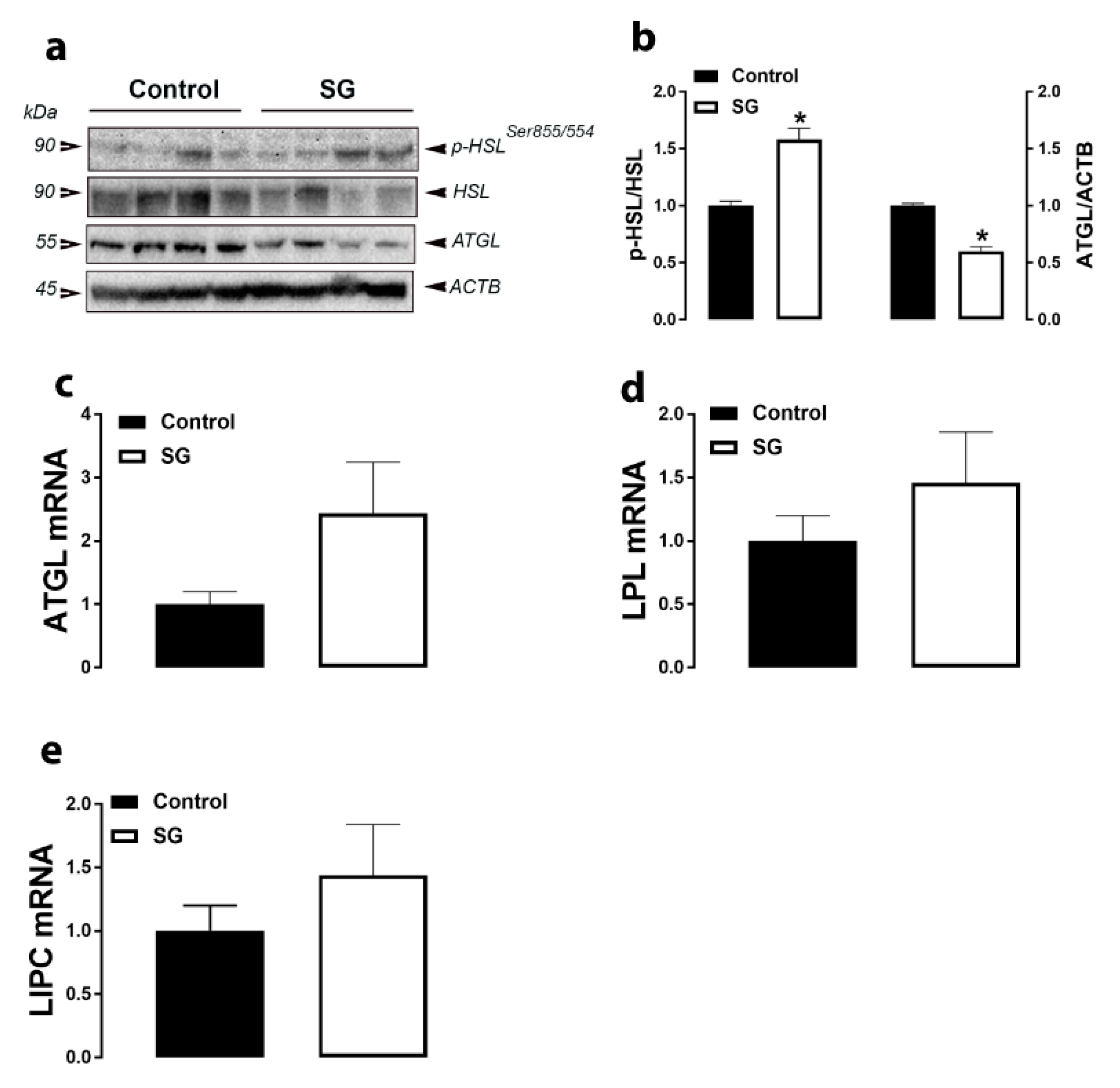


| Parameters b | Experimental Groups c | |||||
|---|---|---|---|---|---|---|
| Control | AVSSL | |||||
| SCD-1 | 1.00 | ± | 0.21 | 1.82 | ± | 0.30 * |
| SREBP1 | 1.00 | ± | 0.11 | 0.79 | ± | 0.10 |
| SREBP2 | 1.00 | ± | 0.26 | 0.46 | ± | 0.05 * |
| SCAP | 1.00 | ± | 0.21 | 0.29 | ± | 0.03 * |
| PPARα | 1.00 | ± | 0.10 | 1.04 | ± | 0.10 |
| PPARγ | 1.00 | ± | 0.14 | 1.00 | ± | 0.09 |
| INSIG2 | 1.00 | ± | 0.14 | 1.00 | ± | 0.14 |
| CD36 | 1.00 | ± | 0.19 | 0.86 | ± | 0.18 |
| AdipoQ | 1.00 | ± | 0.17 | 0.61 | ± | 0.08 * |
| AdipoR1 | 1.00 | ± | 0.11 | 1.01 | ± | 0.11 |
| AdipoR2 | 1.00 | ± | 0.12 | 1.19 | ± | 0.13 |
| NAMPT | 1.00 | ± | 0.11 | 1.30 | ± | 0.15 |
| Parameters b | Experimental Groups c | |||||
|---|---|---|---|---|---|---|
| Control | SG | |||||
| SCD-1 | 1.00 | ± | 0.21 | 1.84 | ± | 0.39 * |
| SREBP1 | 1.00 | ± | 0.11 | 0.80 | ± | 0.08 |
| SREBP2 | 1.00 | ± | 0.26 | 0.55 | ± | 0.08 |
| SCAP | 1.00 | ± | 0.21 | 0.28 | ± | 0.04 * |
| PPARα | 1.00 | ± | 0.10 | 1.16 | ± | 0.15 |
| PPARγ | 1.00 | ± | 0.14 | 1.96 | ± | 0.53 |
| INSIG2 | 1.00 | ± | 0.14 | 0.90 | ± | 0.16 |
| CD36 | 1.00 | ± | 0.19 | 0.72 | ± | 0.15 |
| AdipoQ | 1.00 | ± | 0.17 | 1.48 | ± | 0.34 |
| AdipoR1 | 1.00 | ± | 0.11 | 1.52 | ± | 0.21 * |
| AdipoR2 | 1.00 | ± | 0.12 | 1.30 | ± | 0.11 * |
| NAMPT | 1.00 | ± | 0.11 | 1.43 | ± | 0.25 |
| Parameters b | Experimental Groups c | |||||
|---|---|---|---|---|---|---|
| Control | AVSSL | |||||
| AMPKα1 | 1.00 | ± | 0.27 | 0.97 | ± | 0.10 |
| AMPKα2 | 1.00 | ± | 0.19 | 0.79 | ± | 0.19 |
| AMPKβ1 | 1.00 | ± | 0.05 | 1.66 | ± | 0.20 |
| AMPKβ2 | 1.00 | ± | 0.10 | 0.77 | ± | 0.14 |
| AMPKγ1 | 1.00 | ± | 0.06 | 0.94 | ± | 0.11 |
| AMPKγ2 | 1.00 | ± | 0.18 | 1.45 | ± | 0.21 |
| AMPKγ3 | 1.00 | ± | 0.09 | 0.88 | ± | 0.20 |
| Parameters b | Experimental Groups c | |||||
|---|---|---|---|---|---|---|
| Control | SG | |||||
| AMPKα1 | 1.00 | ± | 0.27 | 0.93 | ± | 0.31 |
| AMPKα2 | 1.00 | ± | 0.19 | 0.93 | ± | 0.15 |
| AMPKβ1 | 1.00 | ± | 0.05 | 1.08 | ± | 0.16 |
| AMPKβ2 | 1.00 | ± | 0.10 | 0.81 | ± | 0.08 |
| AMPKγ1 | 1.00 | ± | 0.06 | 0.89 | ± | 0.05 |
| AMPKγ2 | 1.00 | ± | 0.18 | 1.10 | ± | 0.13 |
| AMPKγ3 | 1.00 | ± | 0.09 | 0.90 | ± | 0.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flees, J.J.; Emami, N.K.; Greene, E.; Ganguly, B.; Dridi, S. Phytogenic Water Additives Improve Broiler Growth Performance via Modulation of Intermediary Metabolism-Related Signaling Pathways. Animals 2021, 11, 750. https://doi.org/10.3390/ani11030750
Flees JJ, Emami NK, Greene E, Ganguly B, Dridi S. Phytogenic Water Additives Improve Broiler Growth Performance via Modulation of Intermediary Metabolism-Related Signaling Pathways. Animals. 2021; 11(3):750. https://doi.org/10.3390/ani11030750
Chicago/Turabian StyleFlees, Joshua J., Nima K. Emami, Elizabeth Greene, Bhaskar Ganguly, and Sami Dridi. 2021. "Phytogenic Water Additives Improve Broiler Growth Performance via Modulation of Intermediary Metabolism-Related Signaling Pathways" Animals 11, no. 3: 750. https://doi.org/10.3390/ani11030750








