Isomer-Specific Effects of cis-9,trans-11- and trans-10,cis-12-CLA on Immune Regulation in Ruminal Epithelial Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Treatments
2.2. RNA Isolation and Real-Time PCR (qPCR) Analysis
2.3. Transcriptome Sequencing
2.4. Data Processing and Analysis
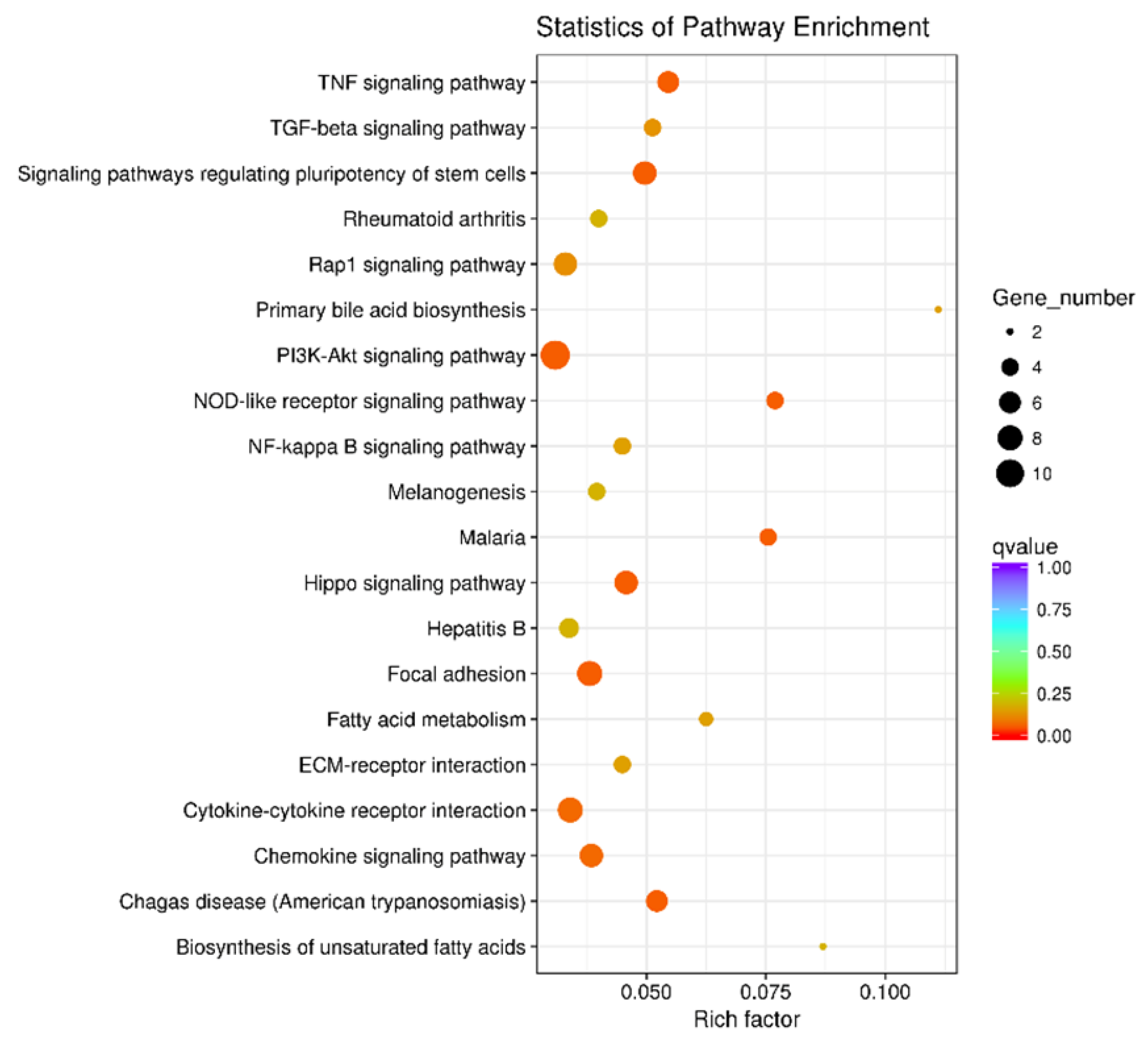
2.5. Functional Analysis of the DEGs
2.6. Statistical Analysis
3. Results
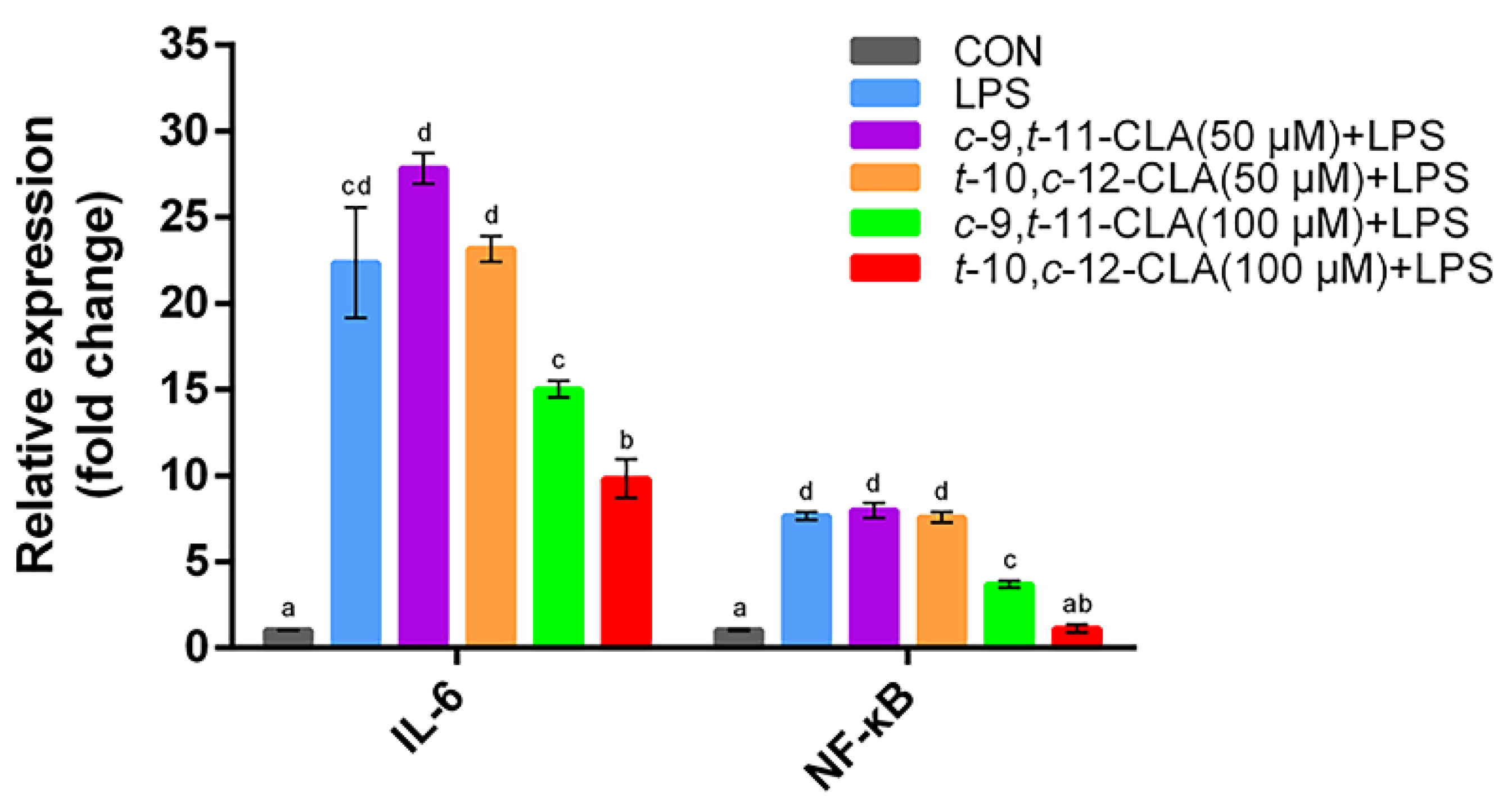
3.1. Optimization of the Concentrations of cis-9,trans-11- and trans-10,cis-12-CLA
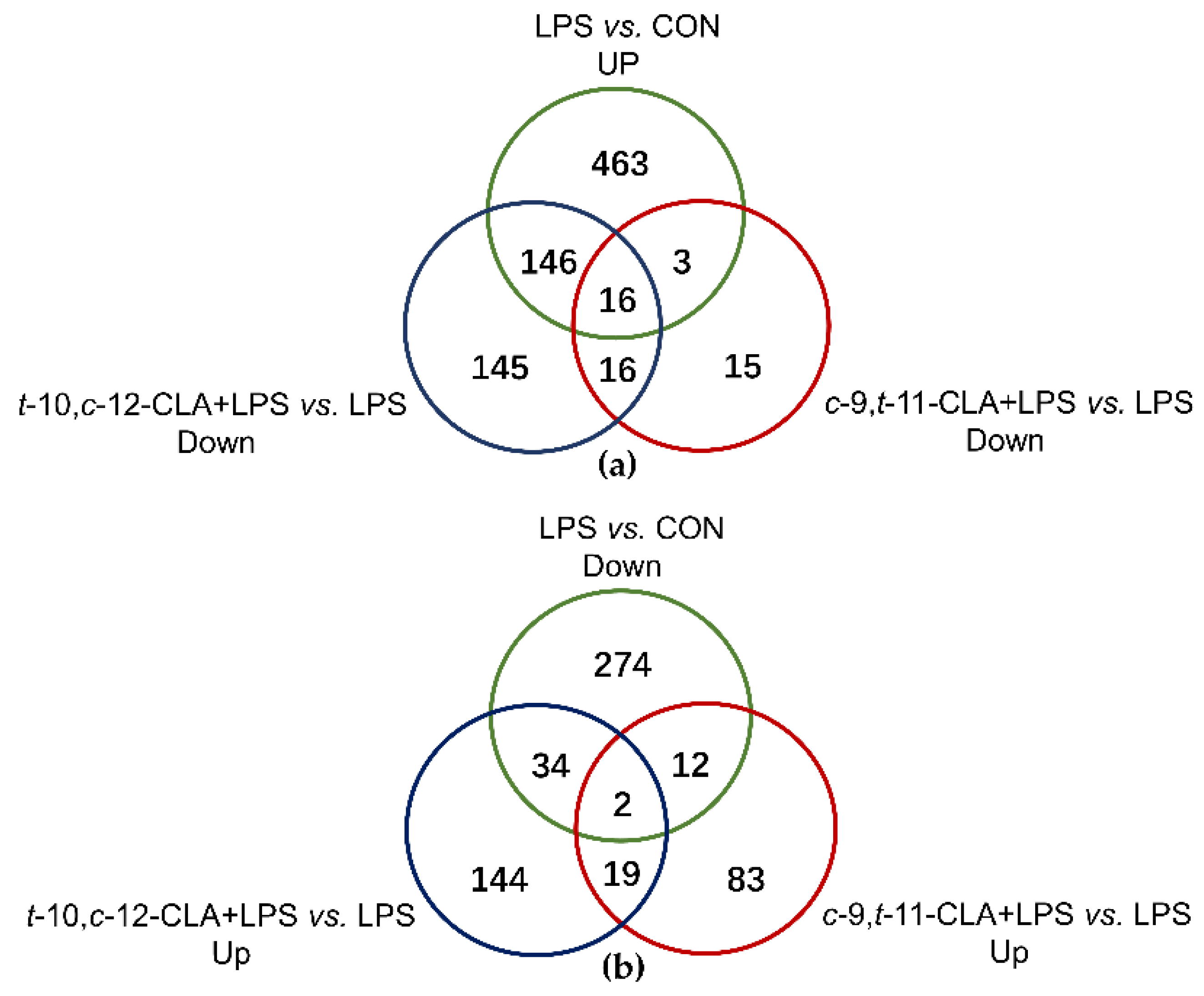
3.2. Overview of the RNA Sequencing Among Different Treatments
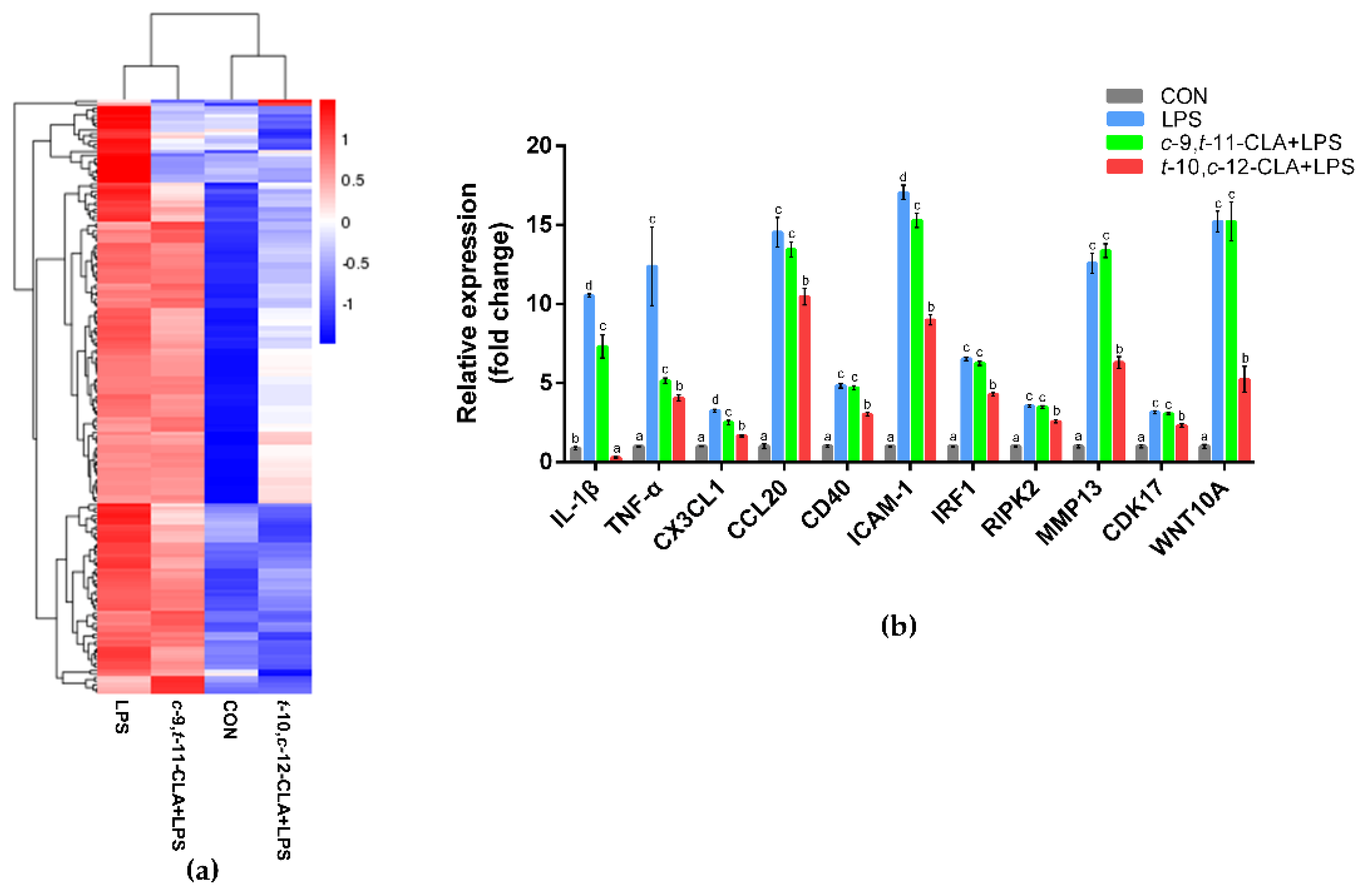
3.3. trans-10,cis-12-CLA Exhibited a Better Effect on Preventing Inflammatory Responses in RECs upon LPS Stimulation than cis-9,trans-11-CLA
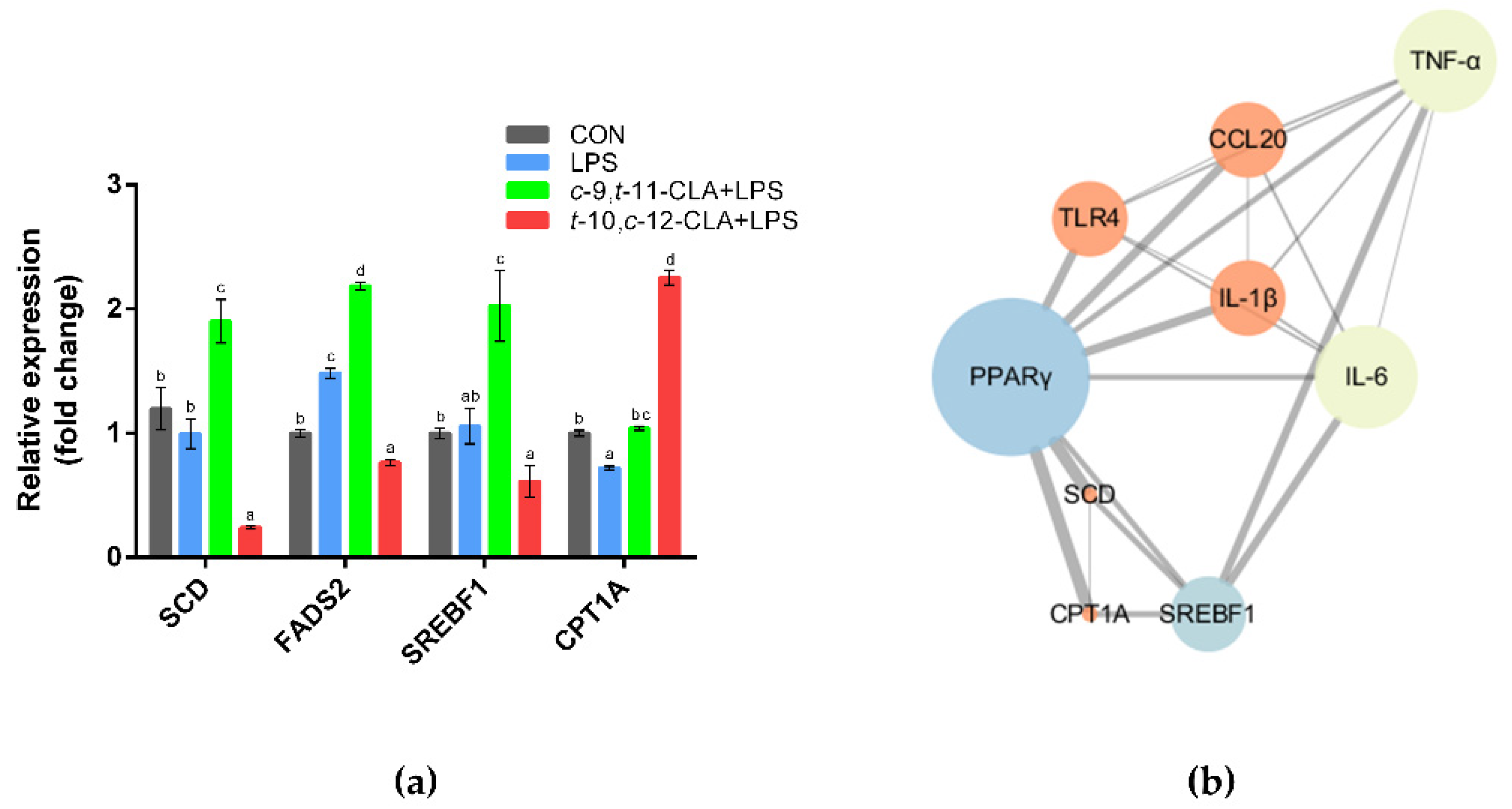
3.4. cis-9,trans-11-CLA and trans-10,cis-12-CLA Affected TLR4 and PPARγ Signaling in an Isomer-Specific Manner
3.5. cis-9,trans-11-CLA and trans-10,cis-12-CLA Affected Adipocytokine Signaling in an Isomer-Specific Manner
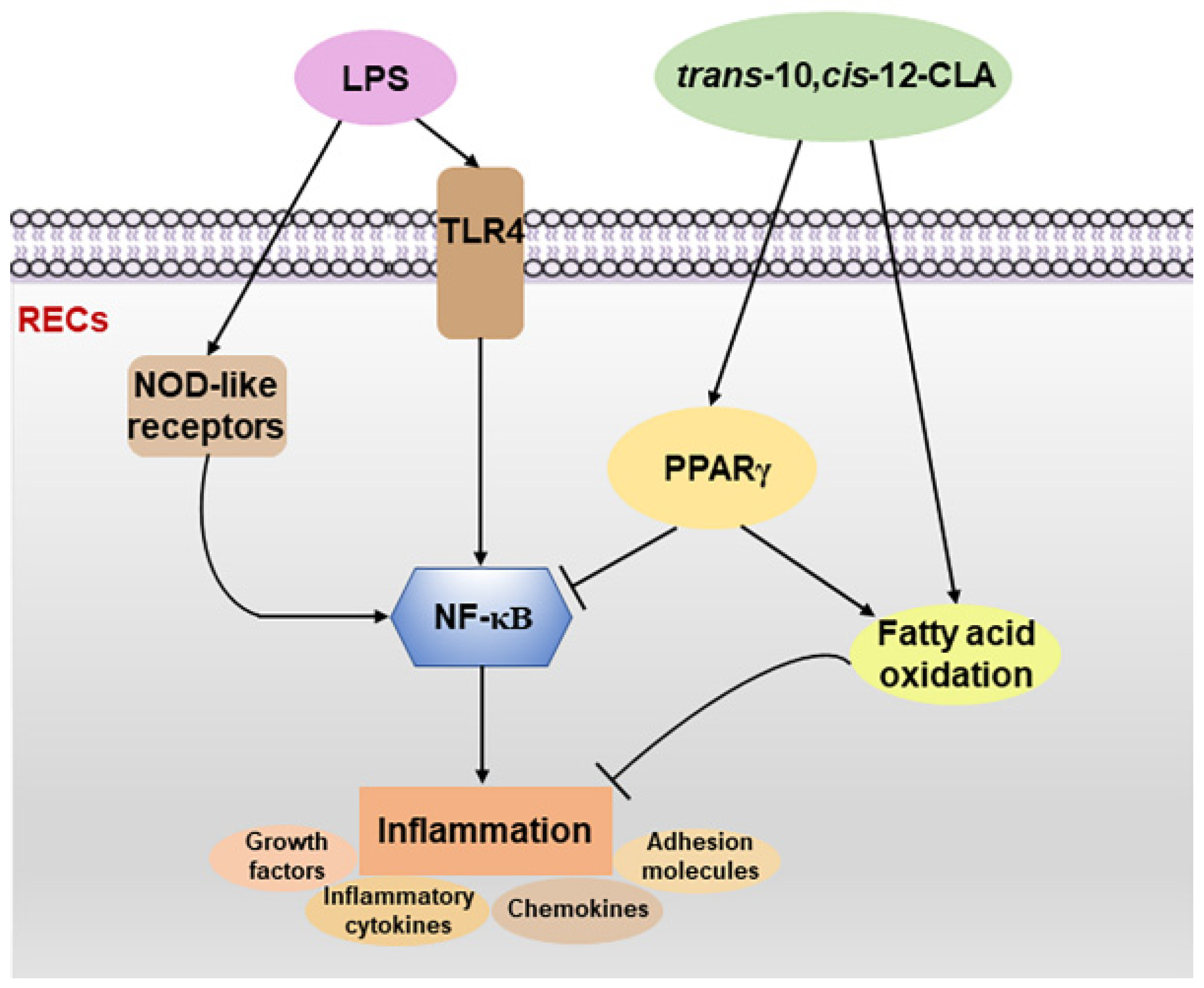
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aschenbach, J.R.; Zebeli, Q.; Patra, A.K.; Greco, G.; Amasheh, S.; Penner, G.B. Symposium review: The importance of the ruminal epithelial barrier for a healthy and productive cow. J. Dairy Sci. 2019, 102, 1866–1882. [Google Scholar] [CrossRef]
- Monteiro, H.F.; Faciola, A.P. Ruminal acidosis, bacterial changes, and lipopolysaccharides. J. Anim. Sci. 2020, 98, skaa248. [Google Scholar] [CrossRef]
- Jiao, J.; Zhou, C.; Guan, L.L.; McSweeney, C.S.; Tang, S.; Wang, M.; Tan, Z. Shifts in host mucosal innate immune function are associated with ruminal microbial succession in supplemental feeding and grazing goats at different ages. Front. Microbiol. 2017, 8, 1655. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Gong, X.; Chen, Y.; Jiang, M.; Yang, T.; Zhao, G. Short-chain fatty acids regulate the immune responses via G protein-coupled receptor 41 in bovine rumen epithelial cells. Front. Immunol. 2019, 10, 2042. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome. Biol. 2019, 20, 172. [Google Scholar] [CrossRef]
- Ferlay, A.; Bernard, L.; Meynadier, A. Malpuech- Brugère. Production of trans and conjugated fatty acids in dairy ruminants and their putative effects on human health: A review. Biochimie 2017, 141, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Bruen, R.; Fitzsimons, S.; Belton, O. Atheroprotective effects of conjugated linoleic acid. Br. J. Clin. Pharmacol. 2017, 83, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Viladomiu, M.; Hontecillas, R.; Bassaganya-Riera, J. Modulation of inflammation and immunity by dietary conjugated linoleic acid. Eur. J. Pharmacol. 2016, 785, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Garibay-Nieto, N.; Queipo-García, G.; Alvarez, F.; Bustos, M.; Villanueva, E.; Ramírez, F.; León, M.; Laresgoiti-Servitje, E.; Duggirala, R.; Macías, T.; et al. Effects of conjugated linoleic acid and metformin on insulin sensitivity in obese children: Randomized clinical trial. J. Clin. Endocrinol. Metab. 2017, 102, 132–140. [Google Scholar] [CrossRef]
- Wang, H.; Liu, T.; Wang, J.; Qi, Y.; Ge, D.; Guan, S. Isomer-specific effects of conjugated linoleic acid on proliferative activity of cultured neural progenitor cells. Mol. Cell. Biochem. 2011, 358, 13–20. [Google Scholar] [CrossRef]
- Ahn, I.S.; Choi, B.H.; Ha, J.H.; Byun, J.M.; Shin, H.G.; Park, K.Y.; Do, M.S. Isomer-specific effect of conjugated linoleic acid on inflammatory adipokines associated with fat accumulation in 3T3-L1 adipocytes. J. Med. Food 2006, 9, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Nicod, N.; Parker, R.S.; Giordano, E.; Maestro, V.; Davalos, A.; Visioli, F. Isomer-specific effects of conjugated linoleic acid on HDL functionality associated with reverse cholesterol transport. J. Nutr. Biochem. 2015, 26, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Della Casa, L.; Rossi, E.; Romanelli, C.; Gibellini, L.; Iannone, A. Effect of diets supplemented with different conjugated linoleic acid (CLA) isomers on protein expression in C57/BL6 mice. Genes. Nutr. 2016, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zhao, W.; Zhang, F.; Song, M.; Liu, F.; Zheng, J.; Ling, M.; Yang, X.; Yang, Q.; He, H.; et al. cis 9, trans 11, but not trans 10, cis 12 CLA isomer, impairs intestinal epithelial barrier function in IPEC-J2 cells and mice through activation of GPR120-[Ca2+] and the MLCK signaling pathway. Food Funct. 2020, 11, 3657–3667. [Google Scholar] [CrossRef] [PubMed]
- Jaudszus, A.; Foerster, M.; Kroegel, C.; Wolf, I.; Jahreis, G. Cis-9,trans-11-CLA exerts anti-inflammatory effects in human bronchial epithelial cells and eosinophils: Comparison to trans-10,cis-12-CLA and to linoleic acid. Biochim. Biophys. Acta 2005, 1737, 111–118. [Google Scholar] [CrossRef]
- Sordillo, L.M. Nutritional strategies to optimize dairy cattle immunity. J. Dairy Sci. 2016, 99, 4967–4982. [Google Scholar] [CrossRef]
- Shokryzadan, P.; Rajion, M.A.; Meng, G.Y.; Boo, L.J.; Ebrahimi, M.; Royan, M.; Sahebi, M.; Azizi, P.; Abiri, R.; Jahromi, M.F. Conjugated linoleic acid: A potent fatty acid linked to animal and human health. Crit. Rev. Food Sci. Nutr. 2017, 57, 2737–2748. [Google Scholar] [CrossRef]
- Zongo, K.; Krishnamoorthy, S.; Moses, J.A.; Yazici, F.; Çon, A.H.; Anandharamakrishnan, C. Total conjugated linoleic acid content of ruminant milk: The world status insights. Food Chem. 2021, 334, 127555. [Google Scholar] [CrossRef]
- Peterson, D.G.; Kelsey, J.A.; Bauman, D.E. Analysis of variation in cis-9, trans-11 conjugated linoleic acid (CLA) in milk fat of dairy cows. J. Dairy Sci. 2002, 85, 2164–2172. [Google Scholar] [CrossRef]
- Yuan, G.; Chen, X.; Li, D. Modulation of peroxisome proliferator-activated receptor gamma (PPAR γ) by conjugated fatty acid in obesity and inflammatory bowel disease. J. Agric. Food Chem. 2015, 63, 1883–1895. [Google Scholar] [CrossRef]
- Mirza, A.Z.; Althagafi, I.I.; Shamshad, H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur. J. Med. Chem. 2019, 166, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Singh, D. Conjugated linoleic acids attenuate FSH- and IGF1-stimulated cell proliferation; IGF1, GATA4, and aromatase expression; and estradiol-17β production in buffalo granulosa cells involving PPARγ, PTEN, and PI3K/Akt. Reproduction 2012, 144, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Lehnen, T.E.; da Silva, M.R.; Camacho, A.; Marcadenti, A.; Lehnen, A.M. A review on effects of conjugated linoleic fatty acid (CLA) upon body composition and energetic metabolism. J. Int. Soc. Sports Nutr. 2015, 12, 36. [Google Scholar] [CrossRef]
- Jung, T.W.; Lee, S.H.; Kim, H.C.; Bang, J.S.; Abd El-Aty, A.M.; Hacımüftüoğlu, A.; Shin, Y.K.; Jeong, J.H. METRNL attenuates lipid-induced inflammation and insulin resistance via AMPK or PPARδ-dependent pathways in skeletal muscle of mice. Exp. Mol. Med. 2018, 50, 122. [Google Scholar] [CrossRef]
- Silva, J.R.; Burger, B.; Kühl, C.M.C.; Candreva, T.; Dos Anjos, M.B.P.; Rodrigues, H.G. Wound healing and Omega-6 fatty acids: From inflammation to repair. Mediat. Inflamm. 2018, 2018, 2503950. [Google Scholar] [CrossRef] [PubMed]
- den Hartigh, L.J. Conjugated linoleic acid effects on cancer, obesity, and atherosclerosis: A review of pre-clinical and human trials with current perspectives. Nutrients 2019, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; McIntosh, M.K. Nutrient regulation: Conjugated linoleic acid’s inflammatory and browning properties in adipose tissue. Annu. Rev. Nutr. 2016, 36, 183–210. [Google Scholar] [CrossRef]
- Rai, M.F.; Tycksen, E.D.; Sandell, L.J.; Brophy, R.H. Advantages of RNA-seq compared to RNA microarrays for transcriptome profiling of anterior cruciate ligament tears. J. Orthop. Res. 2018, 36, 484–497. [Google Scholar] [CrossRef]
- Yang, C.; Lan, W.; Ye, S.; Zhu, B.; Fu, Z.J.F.i.P. Transcriptomic analyses reveal the protective immune regulation of conjugated linoleic acids in sheep ruminal epithelial cells. Front. Physiol. 2020, 11, 588082. [Google Scholar] [CrossRef]
- Zeng, C.; Motta-Ribeiro, G.C.; Hinoshita, T.; Lessa, M.A.; Winkler, T.; Grogg, K.; Kingston, N.M.; Hutchinson, J.N.; Sholl, L.M.; Fang, X.; et al. Lung atelectasis promotes immune and barrier dysfunction as revealed by transcriptome sequencing in female sheep. Anesthesiology 2020, 133, 1060–1076. [Google Scholar] [CrossRef] [PubMed]
- Dipasquale, D.; Basiricò, L.; Morera, P.; Primi, R.; Tröscher, A.; Bernabucci, U. Anti-inflammatory effects of conjugated linoleic acid isomers and essential fatty acids in bovine mammary epithelial cells. Animal 2018, 12, 2108–2114. [Google Scholar] [CrossRef] [PubMed]
- Renner, L.; Kersten, S.; Duevel, A.; Schuberth, H.J.; Dänicke, S. Effects of cis-9,trans-11 and trans-10,cis-12 conjugated linoleic acid, linoleic acid, phytanic acid and the combination of various fatty acids on proliferation and cytokine expression of bovine peripheral blood mononuclear cells. Nutrients 2013, 5, 2667–2683. [Google Scholar] [CrossRef] [PubMed]
- Basiricò, L.; Morera, P.; Dipasquale, D.; Tröscher, A.; Serra, A.; Mele, M.; Bernabucci, U. Conjugated linoleic acid isomers strongly improve the redox status of bovine mammary epithelial cells (BME-UV1). J. Dairy Sci. 2015, 98, 7071–7082. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Woo, S.M.; Kim, W.J.; Lee, B.N.; Nör, J.E.; Min, K.S.; Choi, C.H.; Koh, J.T.; Lee, K.J.; Hwang, Y.C. Simvastatin inhibits the expression of inflammatory cytokines and cell adhesion molecules induced by LPS in human dental pulp cells. Int. Endod. J. 2017, 50, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, Y.; Shao, F. Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol. 2015, 32, 78–83. [Google Scholar] [CrossRef]
- Verhelst, K.; Carpentier, I.; Beyaert, R. Regulation of TNF-induced NF-κB activation by different cytoplasmic ubiquitination events. Cytokine Growth Factor Rev. 2011, 22, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, I.; Erreni, M.; Kamal, M.A.; Porta, C.; Marchesi, F.; Pesce, S.; Pasqualini, F.; Schiarea, S.; Chiabrando, C.; Mantovani, A.; et al. Differential role of Interleukin-1 and Interleukin-6 in K-Ras-driven pancreatic carcinoma undergoing mesenchymal transition. Oncoimmunology 2017, 7, e1388485. [Google Scholar] [CrossRef]
- Numata, T.; Ito, T.; Maeda, T.; Egusa, C.; Tsuboi, R. IL-33 promotes ICAM-1 expression via NF-kB in murine mast cells. Allergol. Int. 2016, 65, 158–165. [Google Scholar] [CrossRef]
- Bhaskar, S.; Sudhakaran, P.R.; Helen, A. Quercetin attenuates atherosclerotic inflammation and adhesion molecule expression by modulating TLR-NF-κB signaling pathway. Cell. Immunol. 2016, 310, 131–140. [Google Scholar] [CrossRef]
- Lee, S.H.; Yamaguchi, K.; Kim, J.S.; Eling, T.E.; Safe, S.; Park, Y.; Baek, S.J. Conjugated linoleic acid stimulates an anti-tumorigenic protein NAG-1 in an isomer specific manner. Carcinogenesis 2006, 27, 972–981. [Google Scholar] [CrossRef]
- Hydbring, P.; Malumbres, M.; Sicinski, P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat. Rev. Mol. Cell Bio. 2016, 17, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, Q.; Tian, H.; Lv, P.; Zhou, C.; Gao, X. Effects of WNT10A on proliferation and differentiation of human dental pulp cells. J. Endod. 2014, 40, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, M.; Chen, Z.; Chen, J.; Chao, Q.; Yin, X.; Quan, M. FTO promotes cell proliferation and migration in esophageal squamous cell carcinoma through up-regulation of MMP13. Exp. Cell Res. 2020, 389, 111894. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.Y.; Shin, S.W.; Ahn, Y.S.; Chung, K.C. Basic fibroblast growth factor-induced activation of novel CREB kinase during the differentiation of immortalized hippocampal cells. J. Biol. Chem. 2001, 276, 13858–13866. [Google Scholar] [CrossRef]
- Song, Y.; Fu, J.; Zhou, M.; Xiao, L.; Feng, X.; Chen, H.; Huang, W. Activated Hippo/Yes-associated protein pathway promotes cell proliferation and anti-apoptosis in endometrial stromal cells of endometriosis. J. Clin. Endocrinol. Metab. 2016, 101, 1552–1561. [Google Scholar] [CrossRef]
- Wei, L.; Yi, Z.; Guo, K.; Long, X. Long noncoding RNA BCAR4 promotes glioma cell proliferation via EGFR/PI3K/AKT signaling pathway. J. Cell. Physiol. 2019, 234, 23608–23617. [Google Scholar] [CrossRef]
- Zhang, Y.; Alexander, P.B.; Wang, X.F. TGF-β family signaling in the control of cell proliferation and survival. Cold. Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef]
- Banerjee, R.; Henson, B.S.; Russo, N.; Tsodikov, A.; D’Silva, N.J. Rap1 mediates galanin receptor 2-induced proliferation and survival in squamous cell carcinoma. Cell. Signal. 2011, 23, 1110–1118. [Google Scholar] [CrossRef]
- Bo, H.; He, J.; Wang, X.; Du, R.; Bei, H.; Chen, J.; Wang, J.; Wu, F.; Zhang, W.; Chen, Q. Danggui Buxue Tang promotes the adhesion and migration of bone marrow stromal cells via the focal adhesion pathway in vitro. J. Ethnopharmacol. 2019, 231, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Landén, N.X.; Li, D.; Ståhle, M. Transition from inflammation to proliferation: A critical step during wound healing. Cell. Mol. Life Sci. 2016, 73, 3861–3885. [Google Scholar] [CrossRef]
- Han, S.W.; Roman, J. Fibronectin induces cell proliferation and inhibits apoptosis in human bronchial epithelial cells: Pro-oncogenic effects mediated by PI3-kinase and NF-kappa B. Oncogene 2006, 25, 4341–4349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Wang, P.; Yan, Y.J.; Li, Y.; Guan, M.W.; Yu, J.J.; Wang, X.D. IL-33 enhances glioma cell migration and invasion by upregulation of MMP2 and MMP9 via the ST2-NF-κB pathway. Oncol. Rep. 2017, 38, 2033–2042. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef]
- Biswas, G.; Bilen, S.; Kono, T.; Sakai, M.; Hikima, J. Inflammatory immune response by lipopolysaccharide-responsive nucleotide binding oligomerization domain (NOD)-like receptors in the Japanese pufferfish (Takifugu rubripes). Dev. Comp. Immunol. 2016, 55, 21–31. [Google Scholar] [CrossRef]
- Jeong, Y.; Kang, M.; Lee, S.; Kim, C.; Kim, J.; Kim, T.; Kim, D.; Kim, D.; Núñez, G.; Park, J. Nod2 and Rip2 contribute to innate immune responses in mouse neutrophils. Immunology 2014, 143, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lan, P.; Hou, X.; Han, Q.; Lu, N.; Li, T.; Jiao, C.; Zhang, J.; Zhang, C.; Tian, Z. HBV inhibits LPS-induced NLRP3 inflammasome activation and IL-1β production via suppressing the NF-κB pathway and ROS production. J. Hepatol. 2017, 66, 693–702. [Google Scholar] [CrossRef]
- Selvanantham, T.; Escalante, N.K.; Cruz Tleugabulova, M.; Fiévé, S.; Girardin, S.E.; Philpott, D.J.; Mallevaey, T. Nod1 and Nod2 enhance TLR-mediated invariant NKT cell activation during bacterial infection. J. Immunol. 2013, 191, 5646–5654. [Google Scholar] [CrossRef] [PubMed]
- Hrdinka, M.; Schlicher, L.; Dai, B.; Pinkas, D.M.; Bufton, J.C.; Picaud, S.; Ward, J.A.; Rogers, C.; Suebsuwong, C.; Nikhar, S.; et al. Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling. EMBO J. 2018, 37, e99372. [Google Scholar] [CrossRef]
- Daynes, R.A.; Jones, D.C. Emerging roles of PPARs in inflammation and immunity. Nat. Rev. Immunol. 2002, 2, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; Draper, E.; Keogh, B.; Rahman, A.; Moloney, A.P.; Mills, K.H.G.; Loscher, C.E.; Roche, H.M. A conjugated linoleic acid-enriched beef diet attenuates lipopolysaccharide-induced inflammation in mice in part through PPARγ-mediated suppression of Toll-like receptor 4. J. Nutr. 2009, 139, 2351–2357. [Google Scholar] [CrossRef] [PubMed]
- Dasu, M.R.; Park, S.; Devaraj, S.; Jialal, I. Pioglitazone inhibits Toll-like receptor expression and activity in human monocytes and db/db mice. Endocrinology 2009, 150, 3457–3464. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.S.; Han, D.S.; Lee, S.H.; Paik, C.H.; Chung, Y.W.; Lee, J.; Hahm, J.S. Attenuation of colonic inflammation by PPARgamma in intestinal epithelial cells: Effect on Toll-like receptor pathway. Dig. Dis. Sci. 2006, 51, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Qu, R.N.; Qu, W. Metformin inhibits LPS-induced inflammatory response in VSMCs by regulating TLR4 and PPAR-γ. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4988–4995. [Google Scholar] [CrossRef] [PubMed]
- Di Cara, F.; Sheshachalam, A.; Braverman, N.E.; Rachubinski, R.A.; Simmonds, A.J. Peroxisome-mediated metabolism is required for immune response to microbial infection. Immunity 2017, 47, 93–106.e107. [Google Scholar] [CrossRef]
- Kalucka, J.; Bierhansl, L.; Conchinha, N.V.; Missiaen, R.; Elia, I.; Brüning, U.; Scheinok, S.; Treps, L.; Cantelmo, A.R.; Dubois, C.; et al. Quiescent endothelial cells upregulate fatty acid β-oxidation for vasculoprotection via redox homeostasis. Cell Metab. 2018, 28, 881–894.e813. [Google Scholar] [CrossRef]
- Raud, B.; Roy, D.G.; Divakaruni, A.S.; Tarasenko, T.N.; Franke, R.; Ma, E.H.; Samborska, B.; Hsieh, W.Y.; Wong, A.H.; Stüve, P.; et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 2018, 28, 504–515.e507. [Google Scholar] [CrossRef]
- Zhao, F.; Xiao, C.; Evans, K.S.; Theivanthiran, T.; DeVito, N.; Holtzhausen, A.; Liu, J.; Liu, X.; Boczkowski, D.; Nair, S.; et al. Paracrine Wnt5a-β-Catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity 2018, 48, 147–160.e147. [Google Scholar] [CrossRef]
- Urrutia, N.; Ying, Y.; Harvatine, K.J. The effect of conjugated linoleic acid, acetate, and their interaction on adipose tissue lipid metabolism in nonlactating cows. J. Dairy Sci. 2017, 100, 5058–5067. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, T.; Li, C.; Wang, J.; Huang, J.; Li, Z. trans-10,cis-12-Conjugated linoleic acid affects expression of lipogenic genes in mammary glands of lactating dairy goats. J. Agric. Food Chem. 2017, 65, 9460–9467. [Google Scholar] [CrossRef] [PubMed]
- Harvatine, K.J.; Perfield, J.W.; Bauman, D.E. Expression of enzymes and key regulators of lipid synthesis is upregulated in adipose tissue during CLA-induced milk fat depression in dairy cows. J. Nutr. 2009, 139, 849–854. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Zhu, B.; Ye, S.; Fu, Z.; Li, J. Isomer-Specific Effects of cis-9,trans-11- and trans-10,cis-12-CLA on Immune Regulation in Ruminal Epithelial Cells. Animals 2021, 11, 1169. https://doi.org/10.3390/ani11041169
Yang C, Zhu B, Ye S, Fu Z, Li J. Isomer-Specific Effects of cis-9,trans-11- and trans-10,cis-12-CLA on Immune Regulation in Ruminal Epithelial Cells. Animals. 2021; 11(4):1169. https://doi.org/10.3390/ani11041169
Chicago/Turabian StyleYang, Chunlei, Binna Zhu, Shijie Ye, Zhengwei Fu, and Jinjun Li. 2021. "Isomer-Specific Effects of cis-9,trans-11- and trans-10,cis-12-CLA on Immune Regulation in Ruminal Epithelial Cells" Animals 11, no. 4: 1169. https://doi.org/10.3390/ani11041169
APA StyleYang, C., Zhu, B., Ye, S., Fu, Z., & Li, J. (2021). Isomer-Specific Effects of cis-9,trans-11- and trans-10,cis-12-CLA on Immune Regulation in Ruminal Epithelial Cells. Animals, 11(4), 1169. https://doi.org/10.3390/ani11041169





