Daytime Dependence of the Activity of the Rat Brain Pyruvate Dehydrogenase Corresponds to the Mitochondrial Sirtuin 3 Level and Acetylation of Brain Proteins, All Regulated by Thiamine Administration Decreasing Phosphorylation of PDHA Ser293
Abstract
:1. Introduction
2. Results
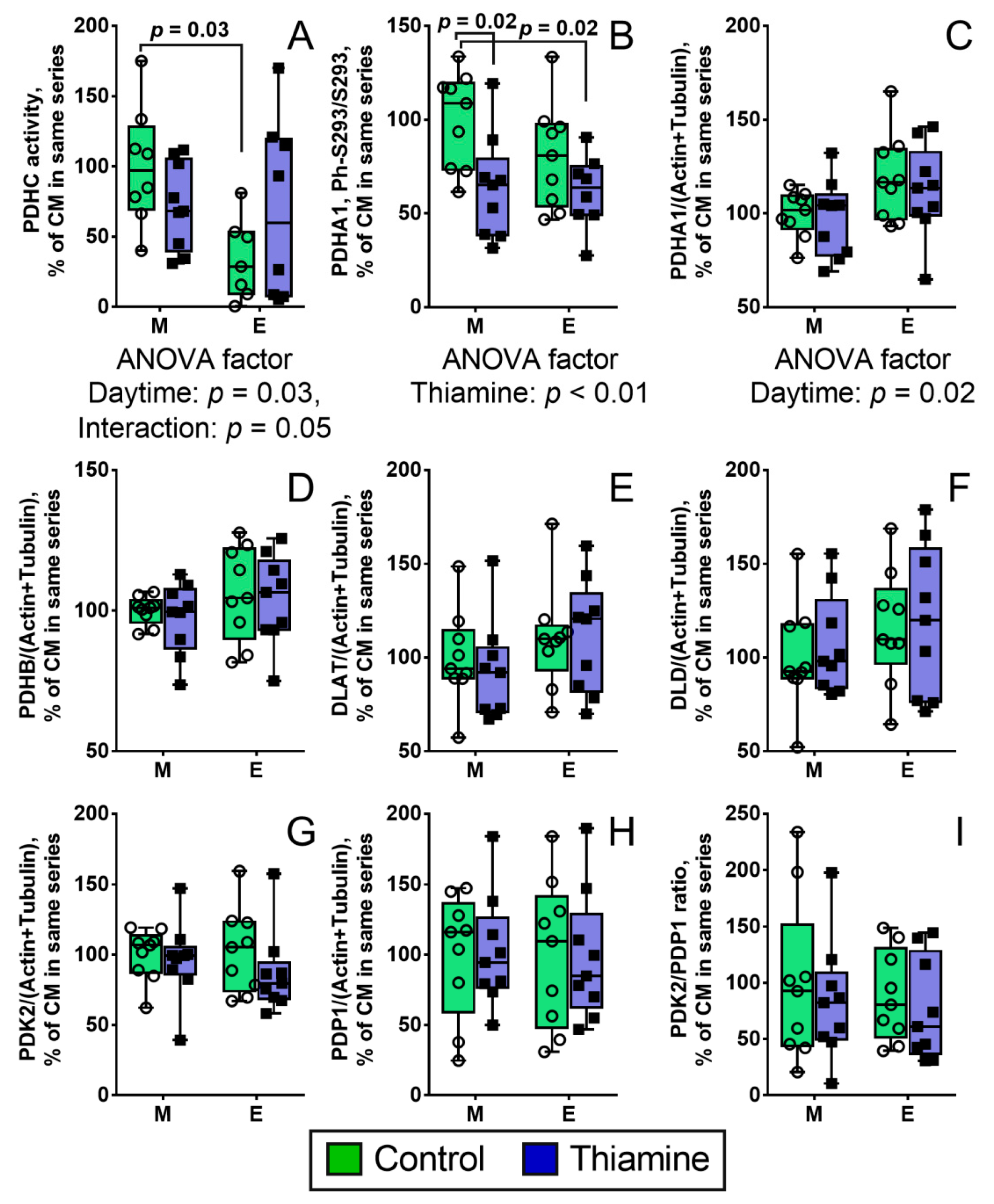
2.1. Thiamine Abrogates the Daytime-Dependent Changes in the Brain PDH Activity and Decreases Ser293 Phosphorylation of PDHA1
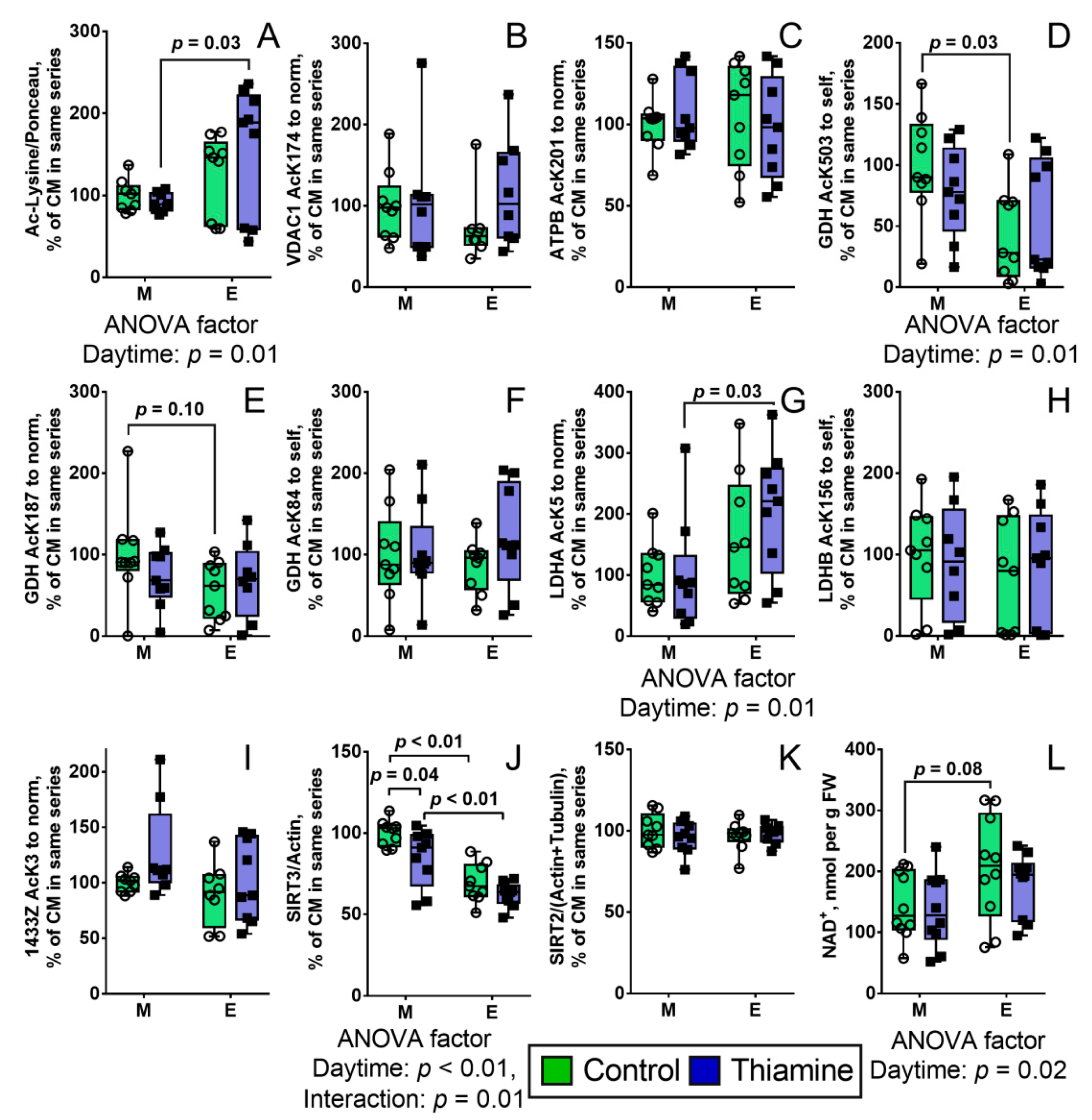
2.2. Diurnal Effect of Thiamine Administration on Protein Acetylation System in the Brain
2.3. Correlation Analysis of Coupled Changes in the PDHC Structural and Functional Parameters and Components of the Brain Proteins Acetylation System
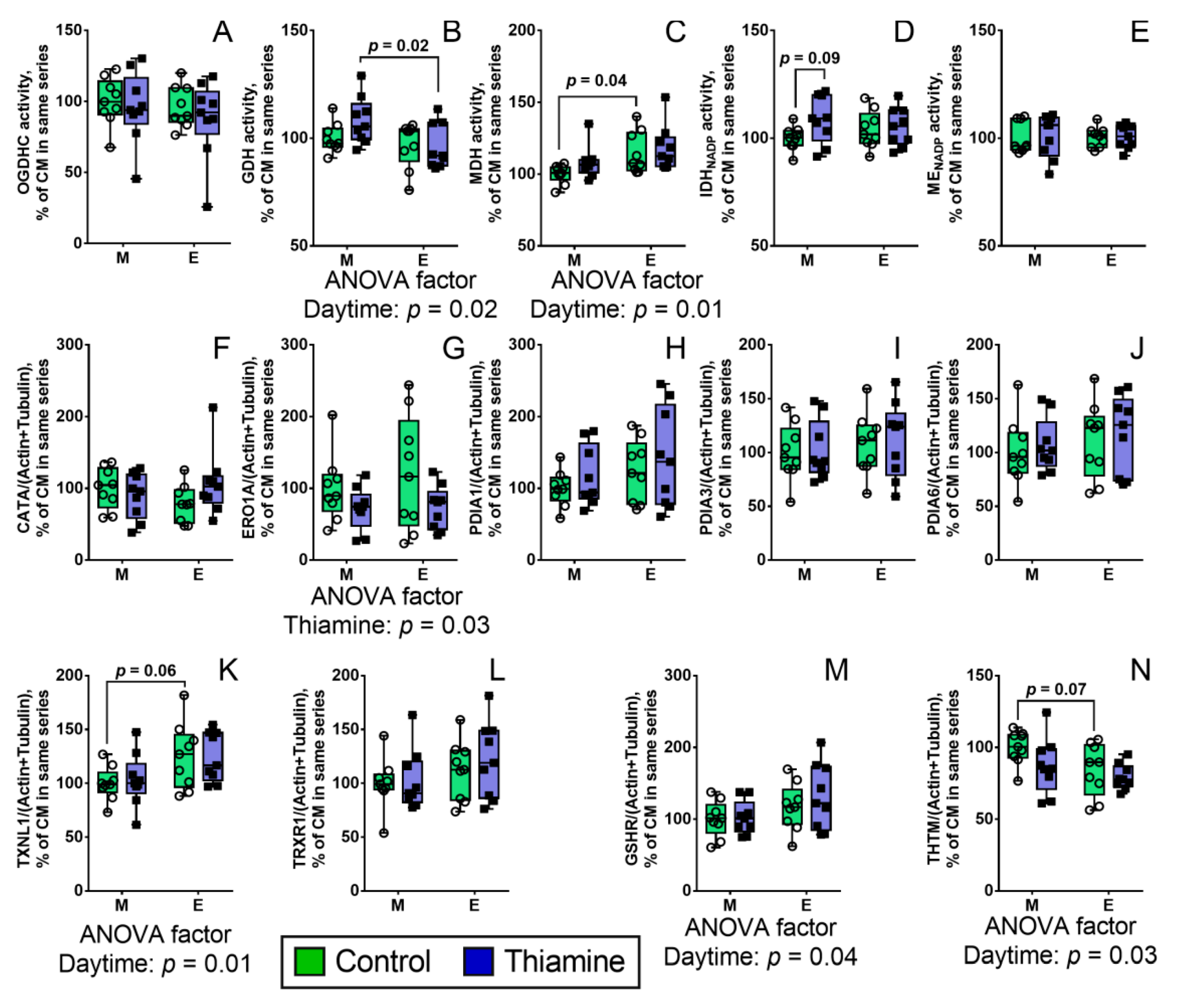
2.4. Daytime- and Thiamine-Dependent Assays of Enzymes of the PDHC- and Redox-Linked Pathways
2.5. Correlation Analysis of the Metabolic Impact of Diurnal Pattern of the Brain Protein Acetylation
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animal Experiments and Thiamine Administration
4.3. Activity Assays
4.4. Analysis of the Peptides by LC-MS/MS
4.5. Quantification of the Protein Levels, PDHA1 Ser293 Phosphorylation, and Acetylation of Other Proteins
4.6. Assay of Total Lysine Acetylation by Western Blotting
4.7. Assay of SIRT3 by Western Blotting
4.8. Preparation of Tissue Extracts and Quantification of NAD+ Levels
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PDH | pyruvate dehydrogenase |
| PDHC | pyruvate dehydrogenase complex |
| OGDH | 2-oxoglutarate dehydrogenase |
| OGDHC | 2-oxoglutarate dehydrogenase complex |
| GDH | glutamate dehydrogenase |
| MDH | malate dehydrogenase |
| IDH | isocitrate dehydrogenase |
| LDH | lactate dehydrogenase |
| ME | malic enzyme |
| ThDP | thiamine diphosphate |
| ANOVA | analysis of variance |
| TBST | Tris-buffered saline with 0.1% Tween-20 |
| MS | mass spectrometry |
| LC-MS/MS | liquid chromatography with tandem mass spectrometry |
| LTQ | linear trap quadropole |
References
- Bunik, V. Vitamin-Dependent Multienzyme Complexes of 2-Oxo Acid Dehydrogenases: Structure, Function, Regulation and Medical Implications; Nova Science Publisher: Hauppauge, NY, USA, 2017; 219p. [Google Scholar]
- Seifert, F.; Ciszak, E.; Korotchkina, L.; Golbik, R.; Spinka, M.; Dominiak, P.; Sidhu, S.; Brauer, J.; Patel, M.S.; Tittmann, K. Phosphorylation of serine 264 impedes active site accessibility in the E1 component of the human pyruvate dehydrogenase multienzyme complex. Biochemistry 2007, 46, 6277–6287. [Google Scholar] [CrossRef]
- Karpova, T.; Danchuk, S.; Kolobova, E.; Popov, K.M. Characterization of the isozymes of pyruvate dehydrogenase phosphatase: Implications for the regulation of pyruvate dehydrogenase activity. Biochim. Biophys. Acta 2003, 1652, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Jha, M.K.; Jeon, S.; Suk, K. Pyruvate Dehydrogenase Kinases in the Nervous System: Their Principal Functions in Neuronal-glial Metabolic Interaction and Neuro-metabolic Disorders. Curr. Neuropharmacol. 2012, 10, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Sutendra, G.; Kinnaird, A.; Dromparis, P.; Paulin, R.; Stenson, T.H.; Haromy, A.; Hashimoto, K.; Zhang, N.; Flaim, E.; Michelakis, E.D. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 2014, 158, 84–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eguchi, K.; Nakayama, K. Prolonged hypoxia decreases nuclear pyruvate dehydrogenase complex and regulates the gene expression. Biochem. Biophys. Res. Commun. 2019, 520, 128–135. [Google Scholar] [CrossRef]
- Lombard, D.B.; Alt, F.W.; Cheng, H.L.; Bunkenborg, J.; Streeper, R.S.; Mostoslavsky, R.; Kim, J.; Yancopoulos, G.; Valenzuela, D.; Murphy, A.; et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007, 27, 8807–8814. [Google Scholar] [CrossRef] [Green Version]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, S.; Moseley, A.C.; Almeda-Valdes, P.; Stromsdorfer, K.L.; Franczyk, M.P.; Okunade, A.L.; Patterson, B.W.; Klein, S.; Yoshino, J. Diurnal Variation in PDK4 Expression Is Associated With Plasma Free Fatty Acid Availability in People. J. Clin. Endocrinol. Metab. 2018, 103, 1068–1076. [Google Scholar] [CrossRef] [Green Version]
- Scrima, R.; Cela, O.; Agriesti, F.; Piccoli, C.; Tataranni, T.; Pacelli, C.; Mazzoccoli, G.; Capitanio, N. Mitochondrial calcium drives clock gene-dependent activation of pyruvate dehydrogenase and of oxidative phosphorylation. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118815. [Google Scholar] [CrossRef]
- Neufeld-Cohen, A.; Robles, M.S.; Aviram, R.; Manella, G.; Adamovich, Y.; Ladeuix, B.; Nir, D.; Rousso-Noori, L.; Kuperman, Y.; Golik, M.; et al. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E1673–E1682. [Google Scholar] [CrossRef] [Green Version]
- Aleshin, V.A.; Mkrtchyan, G.V.; Kaehne, T.; Graf, A.V.; Maslova, M.V.; Bunik, V.I. Diurnal regulation of the function of the rat brain glutamate dehydrogenase by acetylation and its dependence on thiamine administration. J. Neurochem. 2020, 153, 80–102. [Google Scholar] [CrossRef]
- Mauvoisin, D.; Atger, F.; Dayon, L.; Nunez Galindo, A.; Wang, J.; Martin, E.; Da Silva, L.; Montoliu, I.; Collino, S.; Martin, F.P.; et al. Circadian and Feeding Rhythms Orchestrate the Diurnal Liver Acetylome. Cell Rep. 2017, 20, 1729–1743. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Martinez, O.; Mendez, I.; Turrubiate, I.; Valente-Godinez, H.; Perez-Mendoza, M.; Garcia-Tejada, P.; Diaz-Munoz, M. Restricted feeding modulates the daily variations of liver glutamate dehydrogenase activity, expression, and histological location. Exp. Biol. Med. 2017, 242, 945–952. [Google Scholar] [CrossRef] [Green Version]
- Karaca, M.; Frigerio, F.; Migrenne, S.; Martin-Levilain, J.; Skytt, D.M.; Pajecka, K.; Martin-del-Rio, R.; Gruetter, R.; Tamarit-Rodriguez, J.; Waagepetersen, H.S.; et al. GDH-Dependent Glutamate Oxidation in the Brain Dictates Peripheral Energy Substrate Distribution. Cell Rep. 2015, 13, 365–375. [Google Scholar] [CrossRef]
- Kondratova, A.A.; Kondratov, R.V. The circadian clock and pathology of the ageing brain. Nat. Rev. Neurosci. 2012, 13, 325–335. [Google Scholar] [CrossRef]
- Zhang, S.L.; Sehgal, A. Circadian Rhythms and Disease. In Emery and Rimoin’s Principles and Practice of Medical Genetics and Genomics; Academic Press: Cambridge, MA, USA, 2019; pp. 299–314. [Google Scholar] [CrossRef]
- Long, D.M.; Frame, A.K.; Reardon, P.N.; Cumming, R.C.; Hendrix, D.A.; Kretzschmar, D.; Giebultowicz, J.M. Lactate dehydrogenase expression modulates longevity and neurodegeneration in Drosophila melanogaster. Aging 2020, 12, 10041–10058. [Google Scholar] [CrossRef]
- Aleshin, V.A.; Mkrtchyan, G.V.; Bunik, V.I. Mechanisms of the non-coenzyme action of thiamin: Protein targets and medical significance. Biochemistry 2019, 84. [Google Scholar] [CrossRef]
- Mkrtchyan, G.; Aleshin, V.; Parkhomenko, Y.; Kaehne, T.; Di Salvo, M.L.; Parroni, A.; Contestabile, R.; Vovk, A.; Bettendorff, L.; Bunik, V. Molecular mechanisms of the non-coenzyme action of thiamin in brain: Biochemical, structural and pathway analysis. Sci. Rep. 2015, 5, 12583. [Google Scholar] [CrossRef] [Green Version]
- Boyko, A.; Tsepkova, P.; Aleshin, V.; Artiukhov, A.; Mkrtchyan, G.; Ksenofontov, A.; Baratova, L.; Ryabov, S.; Graf, A.; Bunik, V. Severe Spinal Cord Injury in Rats Induces Chronic Changes in the Spinal Cord and Cerebral Cortex Metabolism, Adjusted by Thiamine That Improves Locomotor Performance. Front. Mol. Neurosci. 2021, 14. [Google Scholar] [CrossRef]
- McLure, K.G.; Takagi, M.; Kastan, M.B. NAD+ modulates p53 DNA binding specificity and function. Mol. Cell. Biol. 2004, 24, 9958–9967. [Google Scholar] [CrossRef] [Green Version]
- Bunik, V.I.; Aleshin, V.A. Analysis of the Protein Binding Sites for Thiamin and Its Derivatives to Elucidate the Molecular Mechanisms of the Noncoenzyme Action of Thiamin (Vitamin B1). Stud. Nat. Prod. Chem. 2017, 53, 375–429. [Google Scholar] [CrossRef]
- Parkhomenko Iu, M.; Chernysh, I.; Churilova, T.; Khalmuradov, A.G. Effect of thiamine phosphates on the activity of regulatory enzymes of the pyruvate dehydrogenase complex. Ukr. Biokhimicheskii Zhurnal 1987, 59, 49–54. [Google Scholar]
- Kolobova, E.; Tuganova, A.; Boulatnikov, I.; Popov, K.M. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 2001, 358, 69–77. [Google Scholar] [CrossRef]
- Jonus, H.C.; Byrnes, C.C.; Kim, J.; Valle, M.L.; Bartlett, M.G.; Said, H.M.; Zastre, J.A. Thiamine mimetics sulbutiamine and benfotiamine as a nutraceutical approach to anticancer therapy. Biomed Pharm. 2020, 121, 109648. [Google Scholar] [CrossRef]
- Bunik, V.I.; Aleshin, V.A.; Zhou, X.; Tabakov, V.Y.; Karlsson, A. Activation of Mitochondrial 2-Oxoglutarate Dehydrogenase by Cocarboxylase in Human Lung Adenocarcinoma Cells A549 Is p53/p21-Dependent and Impairs Cellular Redox State, Mimicking the Cisplatin Action. Int. J. Mol. Sci. 2020, 21, 3759. [Google Scholar] [CrossRef]
- Gabriel, B.M.; Zierath, J.R. Zeitgebers of skeletal muscle and implications for metabolic health. J. Physiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Smaaland, R.; Svardal, A.M.; Lote, K.; Ueland, P.M.; Laerum, O.D. Glutathione Content in Human Bone Marrow and Circadian Stage Relation to DNA Synthesis. JNCI J. Natl. Cancer Inst. 1991, 83, 1092–1098. [Google Scholar] [CrossRef]
- Iwata, S.; Ozawa, K.; Shimahara, Y.; Mori, K.; Kobayashi, N.; Kumada, K.; Yamaoka, Y. Diurnal fluctuations of arterial ketone body ratio in normal subjects and patients with liver dysfunction. Gastroenterology 1991, 100, 1371–1378. [Google Scholar] [CrossRef]
- Bunik, V.I.; Fernie, A.R. Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: A cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem. J. 2009, 422, 405–421. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Shan, C.; Kang, H.B.; Elf, S.; Xie, J.; Tucker, M.; Gu, T.L.; Aguiar, M.; Lonning, S.; Chen, H.; et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell 2014, 53, 534–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, S.K.; Hammes, G.G. Elementary steps in the reaction mechanism of the pyruvate dehydrogenase multienzyme complex from Escherichia coli: Kinetics of acetylation and deacetylation. Biochemistry 1980, 19, 4208–4213. [Google Scholar] [CrossRef] [PubMed]
- Khailova, L.S.; Alexandrovitch, O.V.; Severin, S.E. Substrate-dependent inactivation of muscle pyruvate dehydrogenase: Identification of the acetyl-substituted enzyme form. Biochem. Int. 1985, 10, 291–300. [Google Scholar]
- Peek, C.B.; Affinati, A.H.; Ramsey, K.M.; Kuo, H.Y.; Yu, W.; Sena, L.A.; Ilkayeva, O.; Marcheva, B.; Kobayashi, Y.; Omura, C.; et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 2013, 342, 1243417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhang, B.; Tang, J.; Cao, Q.; Wu, Y.; Wu, C.; Guo, J.; Ling, E.A.; Liang, F. Sirtuin 2, a Mammalian Homolog of Yeast Silent Information Regulator-2 Longevity Regulator, Is an Oligodendroglial Protein That Decelerates Cell Differentiation through Deacetylating Tubulin. J. Neurosci. 2007, 27, 2606–2616. [Google Scholar] [CrossRef] [Green Version]
- Esteves, A.R.; Arduíno, D.M.; Silva, D.F.; Viana, S.D.; Pereira, F.C.; Cardoso, S.M. Mitochondrial Metabolism Regulates Microtubule Acetylome and Autophagy Trough Sirtuin-2: Impact for Parkinson’s Disease. Mol. Neurobiol. 2017, 55, 1440–1462. [Google Scholar] [CrossRef]
- Silva, D.F.; Esteves, A.R.; Oliveira, C.R.; Cardoso, S.M. Mitochondrial Metabolism Power SIRT2-Dependent Deficient Traffic Causing Alzheimer’s-Disease Related Pathology. Mol. Neurobiol. 2016, 54, 4021–4040. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Park, S.-H.; Imbesi, M.; Nathan, W.J.; Zou, X.; Zhu, Y.; Jiang, H.; Parisiadou, L.; Gius, D. Loss of NAD-Dependent Protein Deacetylase Sirtuin-2 Alters Mitochondrial Protein Acetylation and Dysregulates Mitophagy. Antioxid. Redox Signal. 2017, 26, 849–863. [Google Scholar] [CrossRef]
- Milev, N.B.; Rhee, S.G.; Reddy, A.B. Cellular Timekeeping: It’s Redox o’Clock. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Soni, S.K.; Basu, P.; Singaravel, M.; Sharma, R.; Pandi-Perumal, S.R.; Cardinali, D.P.; Reiter, R.J. Sirtuins and the circadian clock interplay in cardioprotection: Focus on sirtuin 1. Cell. Mol. Life Sci. CMLS 2021, 78, 2503–2515. [Google Scholar] [CrossRef]
- Vall-Llaura, N.; Reverter-Branchat, G.; Vived, C.; Weertman, N.; Rodriguez-Colman, M.J.; Cabiscol, E. Reversible glutathionylation of Sir2 by monothiol glutaredoxins Grx3/4 regulates stress resistance. Free Radic. Biol. Med. 2016, 96, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.M.; An, A.R.; Park, H.S.; Jang, K.Y.; Moon, W.S.; Kang, M.J.; Lee, Y.C.; Ku, J.H.; Chung, M.J. Combined expression of protein disulfide isomerase and endoplasmic reticulum oxidoreductin 1-alpha is a poor prognostic marker for non-small cell lung cancer. Oncol. Lett. 2018, 16, 5753–5760. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yin, F.; Xu, J.; Zhang, T.; Wang, G.; Mao, M.; Wang, Z.; Sun, W.; Han, J.; Yang, M.; et al. CYT997(Lexibulin) induces apoptosis and autophagy through the activation of mutually reinforced ER stress and ROS in osteosarcoma. J. Exp. Clin. Cancer Res. CR 2019, 38, 44. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Lin, C.; Liu, W.; Huo, Y.; Yang, M.; Jiang, S.H.; Sun, Y.; Hua, R. Endoplasmic Reticulum stress-dependent expression of ERO1L promotes aerobic glycolysis in Pancreatic Cancer. Theranostics 2020, 10, 8400–8414. [Google Scholar] [CrossRef]
- Aleshin, V.A.; Zhou, X.; Krishnan, S.; Karlsson, A.; Bunik, V.I. Interplay Between Thiamine and p53/p21 Axes Affects Antiproliferative Action of Cisplatin in Lung Adenocarcinoma Cells by Changing Metabolism of 2-Oxoglutarate/Glutamate. Front. Genet. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.E.; Hirsch, J.A.; Fonzetti, P.; Jordan, B.D.; Cirio, R.T.; Elder, J. Vitamin B1 (thiamine) and dementia. Ann. N. Y. Acad. Sci. 2016, 1367, 21–30. [Google Scholar] [CrossRef]
- Tsepkova, P.M.; Artiukhov, A.V.; Boyko, A.I.; Aleshin, V.A.; Mkrtchyan, G.V.; Zvyagintseva, M.A.; Ryabov, S.I.; Ksenofontov, A.L.; Baratova, L.A.; Graf, A.V.; et al. Thiamine Induces Long-Term Changes in Amino Acid Profiles and Activities of 2-Oxoglutarate and 2-Oxoadipate Dehydrogenases in Rat Brain. Biochemistry 2017, 82, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Mkrtchyan, G.V.; Ucal, M.; Mullebner, A.; Dumitrescu, S.; Kames, M.; Moldzio, R.; Molcanyi, M.; Schaefer, S.; Weidinger, A.; Schaefer, U.; et al. Thiamine preserves mitochondrial function in a rat model of traumatic brain injury, preventing inactivation of the 2-oxoglutarate dehydrogenase complex. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.E.; Luchsinger, J.A.; Cirio, R.; Chen, H.; Franchino-Elder, J.; Hirsch, J.A.; Bettendorff, L.; Chen, Z.; Flowers, S.A.; Gerber, L.M.; et al. Benfotiamine and Cognitive Decline in Alzheimer’s Disease: Results of a Randomized Placebo-Controlled Phase IIa Clinical Trial. J. Alzheimer’s Dis. 2020, 78, 989–1010. [Google Scholar] [CrossRef]
- Sato, Y.; Kojima, R.; Okumura, M.; Hagiwara, M.; Masui, S.; Maegawa, K.; Saiki, M.; Horibe, T.; Suzuki, M.; Inaba, K. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci. Rep. 2013, 3, 2456. [Google Scholar] [CrossRef]
- Ren, H.; Zhai, W.; Lu, X.; Wang, G. The Cross-Links of Endoplasmic Reticulum Stress, Autophagy, and Neurodegeneration in Parkinson’s Disease. Front. Aging Neurosci. 2021, 13, 691881. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.E.; Huang, H.M. Mitochondrial enzymes and endoplasmic reticulum calcium stores as targets of oxidative stress in neurodegenerative diseases. J. Bioenerg. Biomembr. 2004, 36, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Artiukhov, A.V.; Grabarska, A.; Gumbarewicz, E.; Aleshin, V.A.; Kahne, T.; Obata, T.; Kazantsev, A.V.; Lukashev, N.V.; Stepulak, A.; Fernie, A.R.; et al. Synthetic analogues of 2-oxo acids discriminate metabolic contribution of the 2-oxoglutarate and 2-oxoadipate dehydrogenases in mammalian cells and tissues. Sci. Rep. 2020, 10, 1886. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [Green Version]
- Ksenofontov, A.L.; Boyko, A.I.; Mkrtchyan, G.V.; Tashlitsky, V.N.; Timofeeva, A.V.; Graf, A.V.; Bunik, V.I.; Baratova, L.A. Analysis of Free Amino Acids in Mammalian Brain Extracts. Biochemistry 2017, 82, 1183–1192. [Google Scholar] [CrossRef]
- Artiukhov, A.V.; Pometun, A.A.; Zubanova, S.A.; Tishkov, V.I.; Bunik, V.I. Advantages of formate dehydrogenase reaction for efficient NAD(+) quantification in biological samples. Anal. Biochem. 2020, 603, 113797. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef] [Green Version]
| Thiamine | PDHC Activity | P-Ser293 | PDHA1 | PDHB | DLAT | DLDH | PDK2 | PDP1 | SIRT3 | SIRT2 | AcK Total | AcK174 VDAC1 | AcK201 ATPB | AcK503 GDH | AcK187 GDH | AcK84 GDH | AcK5 LDHA | AcK156 LDHB | AcK3 1433Z | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | ||||||||||||||||||||
| PDHC activity | −0.13 0.61 | −0.06 0.83 | 0.00 1.00 | −0.12 0.65 | −0.28 0.26 | −0.07 0.78 | −0.37 0.14 | 0.00 0.99 | −0.21 0.41 | 0.20 0.44 | 0.32 0.21 | 0.43 0.07 | 0.34 0.16 | 0.36 0.14 | 0.21 0.39 | −0.56 0.02 | 0.18 0.47 | 0.18 0.50 | ||
| P-Ser293 | −0.45 0.06 | 0.43 0.08 | 0.47 0.05 | 0.56 0.02 | 0.61 0.01 | 0.47 0.05 | 0.37 0.13 | 0.00 1.00 | 0.51 0.03 | −0.74 0.00 | −0.48 0.05 | −0.46 0.06 | −0.33 0.18 | −0.23 0.37 | −0.31 0.21 | 0.06 0.82 | 0.33 0.19 | −0.09 0.75 | ||
| PDHA1 | −0.17 0.49 | 0.52 0.03 | 0.88 0.00 | 0.25 0.32 | 0.26 0.30 | 0.88 0.00 | 0.19 0.44 | −0.08 0.75 | 0.56 0.02 | −0.57 0.01 | −0.52 0.03 | 0.23 0.36 | −0.26 0.31 | 0.14 0.58 | −0.12 0.63 | −0.32 0.19 | 0.14 0.58 | −0.55 0.03 | ||
| PDHB | −0.31 0.22 | 0.68 0.00 | 0.92 0.00 | 0.27 0.28 | 0.35 0.15 | 0.94 0.00 | 0.30 0.22 | 0.16 0.54 | 0.75 0.00 | −0.56 0.02 | −0.58 0.01 | 0.16 0.52 | −0.39 0.11 | 0.00 0.99 | −0.30 0.22 | −0.46 0.06 | 0.11 0.67 | −0.43 0.09 | ||
| DLAT | −0.50 0.03 | 0.24 0.34 | 0.27 0.28 | 0.32 0.20 | 0.81 0.00 | 0.42 0.08 | 0.55 0.02 | 0.05 0.84 | 0.58 0.01 | −0.49 0.04 | −0.29 0.27 | −0.32 0.20 | −0.54 0.02 | −0.44 0.07 | −0.65 0.00 | 0.09 0.72 | 0.34 0.17 | −0.16 0.55 | ||
| DLD | −0.51 0.03 | 0.37 0.13 | 0.36 0.14 | 0.39 0.11 | 0.97 0.00 | 0.40 0.10 | 0.76 0.00 | 0.27 0.28 | 0.64 0.00 | −0.62 0.01 | −0.43 0.08 | −0.26 0.29 | −0.70 0.00 | −0.63 0.00 | −0.77 0.00 | 0.14 0.57 | 0.41 0.09 | 0.11 0.68 | ||
| PDK2 | −0.17 0.51 | 0.61 0.01 | 0.95 0.00 | 0.94 0.00 | 0.33 0.18 | 0.40 0.10 | 0.37 0.14 | 0.14 0.59 | 0.81 0.00 | −0.56 0.02 | −0.60 0.01 | 0.07 0.77 | −0.36 0.14 | −0.06 0.82 | −0.31 0.21 | −0.38 0.12 | 0.10 0.70 | −0.40 0.13 | ||
| PDP1 | −0.42 0.08 | 0.36 0.14 | 0.41 0.09 | 0.48 0.04 | 0.87 0.00 | 0.87 0.00 | 0.47 0.05 | 0.42 0.09 | 0.71 0.00 | −0.28 0.25 | −0.47 0.06 | −0.40 0.10 | −0.67 0.00 | −0.60 0.01 | −0.69 0.00 | 0.11 0.66 | 0.27 0.29 | 0.10 0.71 | ||
| SIRT3 | 0.42 0.08 | −0.03 0.90 | −0.09 0.72 | −0.03 0.90 | 0.04 0.87 | −0.05 0.85 | 0.09 0.73 | −0.11 0.68 | 0.31 0.21 | −0.11 0.65 | −0.42 0.10 | 0.08 0.75 | −0.17 0.50 | −0.39 0.11 | −0.20 0.43 | −0.25 0.31 | 0.02 0.95 | 0.43 0.10 | ||
| SIRT2 | −0.45 0.06 | 0.76 0.00 | 0.64 0.00 | 0.83 0.00 | 0.31 0.22 | 0.38 0.12 | 0.76 0.00 | 0.44 0.06 | 0.08 0.75 | −0.43 0.07 | −0.52 0.03 | −0.19 0.45 | −0.64 0.00 | −0.38 0.11 | −0.65 0.00 | −0.20 0.41 | 0.12 0.63 | −0.26 0.34 | ||
| AcK total | 0.72 0.00 | −0.64 0.00 | −0.44 0.07 | −0.54 0.02 | −0.44 0.07 | −0.59 0.01 | −0.43 0.07 | −0.49 0.04 | 0.43 0.08 | −0.64 0.00 | 0.73 0.00 | 0.05 0.84 | 0.28 0.27 | 0.08 0.76 | 0.23 0.35 | 0.10 0.68 | −0.28 0.26 | 0.15 0.59 | ||
| AcK174 VDAC1 | 0.61 0.01 | −0.69 0.00 | −0.67 0.00 | −0.71 0.00 | −0.29 0.26 | −0.46 0.07 | −0.60 0.01 | −0.38 0.13 | 0.37 0.14 | −0.70 0.00 | 0.86 0.00 | −0.05 0.85 | 0.24 0.35 | −0.07 0.80 | 0.12 0.64 | −0.01 0.97 | −0.15 0.58 | 0.11 0.69 | ||
| AcK201 ATPB | 0.44 0.07 | −0.10 0.68 | 0.17 0.51 | 0.13 0.60 | −0.10 0.699 | −0.17 0.50 | 0.21 0.40 | 0.04 0.86 | 0.22 0.37 | 0.00 0.99 | 0.49 0.04 | 0.31 0.23 | 0.27 0.28 | 0.50 0.04 | 0.29 0.24 | −0.27 0.28 | 0.08 0.75 | −0.02 0.93 | ||
| AcK503 GDH | 0.62 0.01 | −0.47 0.05 | −0.27 0.28 | −0.44 0.07 | −0.66 0.00 | −0.69 0.00 | −0.39 0.11 | −0.79 0.00 | 0.15 0.56 | −0.67 0.00 | 0.61 0.01 | 0.47 0.05 | 0.23 0.36 | 0.68 0.00 | 0.94 0.00 | 0.05 0.85 | −0.13 0.60 | 0.19 0.49 | ||
| AcK187 GDH | 0.64 0.00 | −0.42 0.08 | −0.14 0.59 | −0.30 0.22 | −0.53 0.02 | −0.63 0.01 | −0.18 0.48 | −0.66 0.00 | 0.42 0.08 | −0.52 0.03 | 0.80 0.00 | 0.58 0.01 | 0.46 0.05 | 0.82 0.00 | 0.76 0.00 | −0.07 0.77 | −0.02 0.93 | −0.17 0.54 | ||
| AcK84 GDH | 0.52 0.03 | −0.50 0.04 | −0.22 0.38 | −0.36 0.14 | −0.69 0.00 | −0.73 0.00 | −0.32 0.20 | −0.77 0.00 | 0.04 0.89 | −0.52 0.03 | 0.56 0.02 | 0.51 0.03 | 0.17 0.51 | 0.82 0.00 | 0.71 0.00 | 0.05 0.85 | −0.23 0.35 | 0.07 0.79 | ||
| AcK5 LDHA | −0.10 0.68 | 0.17 0.51 | −0.02 0.92 | −0.15 0.54 | −0.21 0.41 | −0.11 0.67 | −0.11 0.68 | −0.28 0.26 | −0.16 0.52 | −0.23 0.35 | −0.08 0.75 | −0.15 0.55 | 0.02 0.94 | 0.36 0.15 | 0.27 0.28 | 0.10 0.69 | 0.17 0.49 | 0.02 0.93 | ||
| AcK156 LDHB | −0.06 0.82 | 0.38 0.12 | 0.07 0.78 | 0.20 0.43 | −0.09 0.73 | −0.04 0.89 | 0.13 0.61 | −0.06 0.80 | 0.11 0.65 | 0.10 0.70 | −0.24 0.34 | −0.10 0.70 | −0.08 0.75 | 0.11 0.66 | 0.03 0.91 | 0.00 0.99 | 0.39 0.11 | 0.03 0.91 | ||
| AcK3 1433Z | −0.44 0.09 | −0.01 0.97 | −0.24 0.37 | −0.09 0.74 | −0.11 0.69 | −0.19 0.47 | −0.20 0.46 | −0.11 0.68 | −0.08 0.77 | 0.11 0.69 | −0.17 0.52 | −0.11 0.68 | −0.48 0.06 | −0.27 0.32 | −0.24 0.37 | −0.06 0.84 | −0.37 0.16 | −0.05 0.84 | ||
| Thiamine | AcK Total | SIRT3 | SIRT2 | PDHC act | OGDHC act | IDH act | GDH act | MDH act | ME act | CATA | THTM | ERO1A | PDIA1 | PDIA3 | PDIA6 | GSHR | TRXR1 | TXNL1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | |||||||||||||||||||
| AcK | −0.11 0.65 | −0.43 0.07 | 0.20 0.44 | 0.20 0.42 | −0.06 0.83 | −0.36 0.14 | −0.55 0.02 | −0.31 0.21 | 0.53 0.03 | −0.42 0.08 | −0.80 0.00 | −0.27 0.27 | −0.38 0.12 | −0.62 0.01 | −0.34 0.17 | −0.42 0.08 | 0.42 0.08 | ||
| SIRT3 | 0.43 0.08 | 0.31 0.21 | 0.00 0.99 | 0.06 0.83 | 0.10 0.71 | 0.09 0.73 | −0.23 0.36 | 0.13 0.61 | −0.04 0.89 | 0.42 0.09 | −0.04 0.88 | 0.22 0.38 | 0.41 0.09 | 0.24 0.33 | 0.18 0.48 | 0.28 0.26 | 0.25 0.32 | ||
| SIRT2 | −0.64 0.00 | 0.08 0.75 | −0.21 0.41 | −0.16 0.53 | 0.12 0.63 | −0.02 0.94 | 0.28 0.26 | 0.62 0.01 | −0.23 0.36 | 0.92 0.00 | 0.64 0.00 | 0.09 0.72 | 0.53 0.02 | 0.62 0.01 | 0.49 0.04 | 0.67 0.00 | 0.22 0.38 | ||
| PDHC act | 0.72 0.00 | 0.42 0.08 | −0.45 0.06 | 0.09 0.73 | −0.27 0.27 | 0.28 0.26 | −0.25 0.32 | −0.47 0.05 | 0.49 0.04 | −0.24 0.34 | −0.08 0.75 | −0.25 0.31 | −0.26 0.29 | −0.23 0.36 | −0.46 0.05 | −0.30 0.23 | −0.29 0.24 | ||
| OGDHC act | 0.72 0.00 | 0.18 0.46 | −0.66 0.00 | 0.29 0.24 | 0.24 0.34 | −0.10 0.69 | −0.50 0.04 | −0.52 0.03 | 0.15 0.56 | −0.29 0.24 | −0.31 0.22 | 0.42 0.08 | 0.14 0.57 | −0.15 0.54 | −0.05 0.86 | −0.07 0.79 | 0.19 0.44 | ||
| IDH act | 0.22 0.37 | −0.09 0.72 | −0.53 0.02 | 0.33 0.18 | 0.15 0.56 | 0.09 0.73 | −0.14 0.57 | −0.04 0.87 | −0.06 0.81 | 0.05 0.83 | −0.18 0.48 | 0.14 0.59 | 0.04 0.88 | −0.10 0.68 | 0.09 0.73 | 0.00 1.00 | −0.03 0.89 | ||
| GDH act | 0.07 0.78 | 0.31 0.21 | 0.03 0.92 | 0.17 0.50 | −0.05 0.85 | 0.22 0.38 | 0.00 0.99 | −0.01 0.98 | −0.12 0.63 | 0.02 0.95 | 0.17 0.50 | 0.03 0.92 | −0.22 0.37 | −0.04 0.87 | −0.32 0.19 | −0.30 0.22 | −0.63 0.01 | ||
| MDH act | −0.56 0.02 | −0.44 0.06 | 0.52 0.03 | −0.57 0.01 | −0.47 0.05 | −0.28 0.26 | 0.15 0.54 | 0.66 0.00 | −0.28 0.26 | 0.26 0.30 | 0.71 0.00 | 0.03 0.92 | 0.28 0.26 | 0.59 0.01 | 0.44 0.07 | 0.42 0.08 | −0.31 0.21 | ||
| ME act | −0.58 0.01 | −0.26 0.30 | 0.36 0.15 | −0.65 0.00 | −0.19 0.44 | −0.31 0.21 | −0.49 0.04 | 0.40 0.10 | −0.15 0.55 | 0.69 0.00 | 0.51 0.03 | −0.03 0.91 | 0.31 0.22 | 0.48 0.04 | 0.45 0.06 | 0.44 0.07 | −0.05 0.85 | ||
| CATA | 0.63 0.00 | 0.62 0.01 | −0.31 0.21 | 0.34 0.17 | 0.62 0.01 | 0.17 0.51 | 0.51 0.03 | −0.35 0.16 | −0.42 0.09 | −0.31 0.21 | −0.39 0.11 | −0.45 0.06 | −0.35 0.15 | −0.52 0.03 | −0.43 0.07 | −0.38 0.12 | 0.07 0.78 | ||
| THTM | −0.43 0.08 | 0.31 0.21 | 0.90 0.00 | −0.30 0.23 | −0.52 0.03 | −0.36 0.15 | 0.02 0.93 | 0.28 0.27 | 0.25 0.32 | −0.14 0.58 | 0.57 0.01 | 0.03 0.89 | 0.47 0.05 | 0.56 0.01 | 0.43 0.08 | 0.58 0.01 | 0.17 0.51 | ||
| ERO1A | −0.50 0.03 | −0.03 0.92 | 0.62 0.01 | −0.28 0.26 | −0.46 0.06 | −0.39 0.11 | 0.23 0.36 | 0.62 0.01 | 0.32 0.19 | −0.22 0.39 | 0.48 0.04 | 0.26 0.30 | 0.55 0.02 | 0.80 0.00 | 0.52 0.03 | 0.63 0.01 | −0.25 0.32 | ||
| PDIA1 | −0.53 0.02 | −0.04 0.89 | 0.14 0.59 | −0.53 0.02 | −0.25 0.33 | −0.05 0.85 | 0.10 0.69 | 0.31 0.22 | 0.54 0.02 | −0.12 0.63 | 0.15 0.54 | 0.48 0.04 | 0.76 0.00 | 0.55 0.02 | 0.69 0.00 | 0.56 0.02 | 0.02 0.93 | ||
| PDIA3 | −0.40 0.10 | 0.14 0.58 | 0.20 0.42 | −0.39 0.11 | −0.21 0.39 | −0.06 0.80 | −0.03 0.89 | 0.25 0.32 | 0.50 0.03 | −0.13 0.60 | 0.25 0.31 | 0.58 0.01 | 0.87 0.00 | 0.85 0.00 | 0.90 0.00 | 0.92 0.00 | 0.28 0.26 | ||
| PDIA6 | −0.53 0.02 | 0.01 0.97 | 0.43 0.07 | −0.48 0.04 | −0.37 0.13 | −0.28 0.27 | 0.25 0.31 | 0.53 0.02 | 0.45 0.06 | −0.20 0.43 | 0.38 0.12 | 0.82 0.00 | 0.81 0.00 | 0.83 0.00 | 0.83 0.00 | 0.90 0.00 | 0.01 0.98 | ||
| GSHR | −0.46 0.06 | 0.00 0.99 | 0.17 0.49 | −0.28 0.27 | −0.41 0.09 | 0.00 1.00 | −0.18 0.48 | 0.18 0.47 | 0.46 0.05 | −0.45 0.06 | 0.21 0.39 | 0.44 0.07 | 0.78 0.00 | 0.89 0.00 | 0.71 0.00 | 0.93 0.00 | 0.22 0.38 | ||
| TRXR1 | −0.57 0.01 | 0.00 0.99 | 0.37 0.13 | −0.56 0.02 | −0.30 0.23 | −0.22 0.39 | −0.03 0.92 | 0.45 0.06 | 0.63 0.00 | −0.22 0.38 | 0.35 0.16 | 0.67 0.00 | 0.91 0.00 | 0.94 0.00 | 0.90 0.00 | 0.82 0.00 | 0.27 0.27 | ||
| TXNL1 | 0.05 0.85 | −0.23 0.36 | −0.21 0.39 | −0.16 0.53 | 0.09 0.72 | 0.22 0.37 | −0.71 0.00 | −0.12 0.63 | 0.40 0.10 | −0.19 0.46 | −0.04 0.87 | −0.23 0.37 | 0.21 0.41 | 0.28 0.26 | −0.10 0.69 | 0.32 0.20 | 0.20 0.42 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleshin, V.A.; Artiukhov, A.V.; Kaehne, T.; Graf, A.V.; Bunik, V.I. Daytime Dependence of the Activity of the Rat Brain Pyruvate Dehydrogenase Corresponds to the Mitochondrial Sirtuin 3 Level and Acetylation of Brain Proteins, All Regulated by Thiamine Administration Decreasing Phosphorylation of PDHA Ser293. Int. J. Mol. Sci. 2021, 22, 8006. https://doi.org/10.3390/ijms22158006
Aleshin VA, Artiukhov AV, Kaehne T, Graf AV, Bunik VI. Daytime Dependence of the Activity of the Rat Brain Pyruvate Dehydrogenase Corresponds to the Mitochondrial Sirtuin 3 Level and Acetylation of Brain Proteins, All Regulated by Thiamine Administration Decreasing Phosphorylation of PDHA Ser293. International Journal of Molecular Sciences. 2021; 22(15):8006. https://doi.org/10.3390/ijms22158006
Chicago/Turabian StyleAleshin, Vasily A., Artem V. Artiukhov, Thilo Kaehne, Anastasia V. Graf, and Victoria I. Bunik. 2021. "Daytime Dependence of the Activity of the Rat Brain Pyruvate Dehydrogenase Corresponds to the Mitochondrial Sirtuin 3 Level and Acetylation of Brain Proteins, All Regulated by Thiamine Administration Decreasing Phosphorylation of PDHA Ser293" International Journal of Molecular Sciences 22, no. 15: 8006. https://doi.org/10.3390/ijms22158006
APA StyleAleshin, V. A., Artiukhov, A. V., Kaehne, T., Graf, A. V., & Bunik, V. I. (2021). Daytime Dependence of the Activity of the Rat Brain Pyruvate Dehydrogenase Corresponds to the Mitochondrial Sirtuin 3 Level and Acetylation of Brain Proteins, All Regulated by Thiamine Administration Decreasing Phosphorylation of PDHA Ser293. International Journal of Molecular Sciences, 22(15), 8006. https://doi.org/10.3390/ijms22158006







