Enhancement of Transgene Expression by Mild Hypothermia Is Promoter Dependent in HEK293 Cells
Abstract
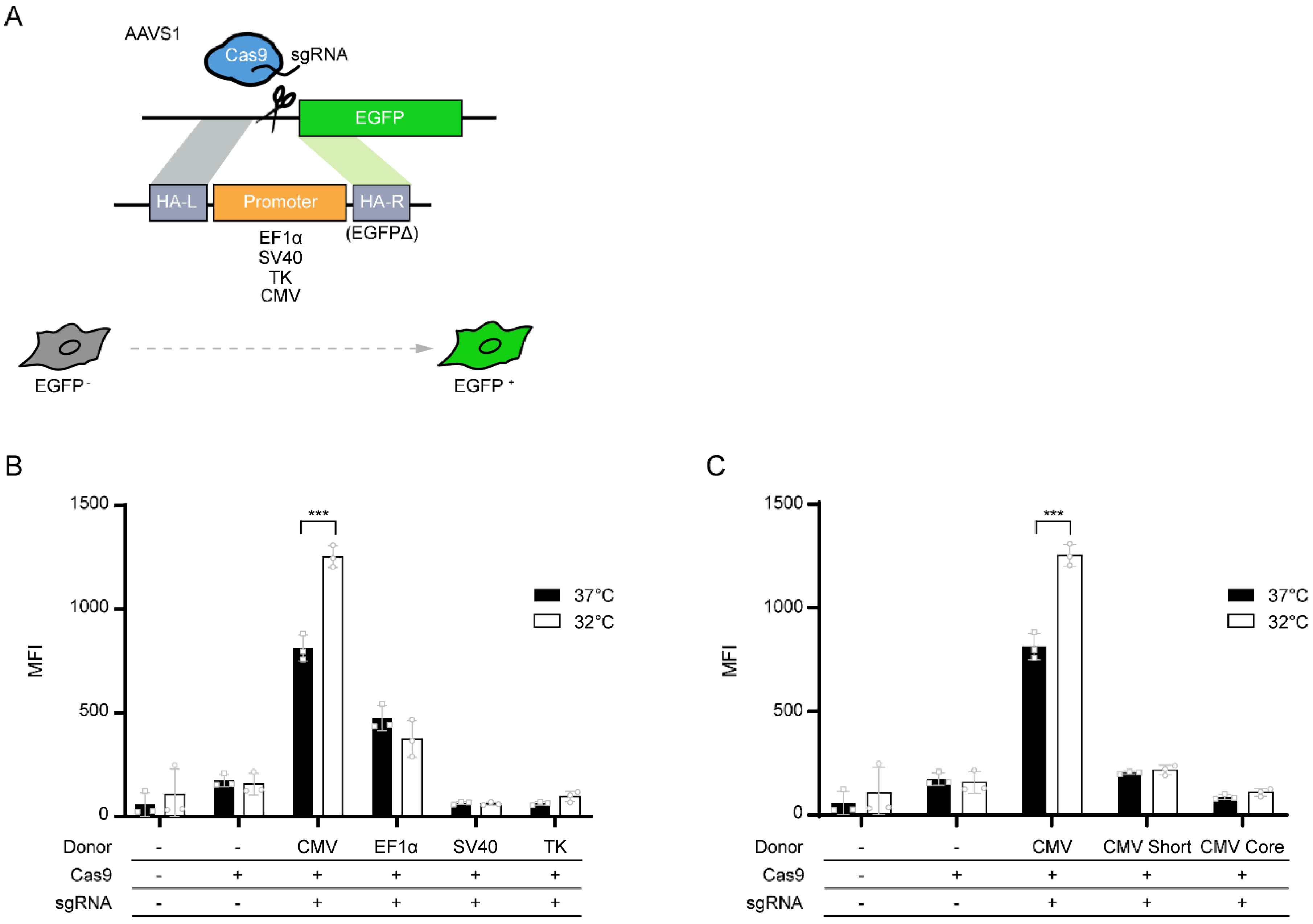
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Plasmids and Transfection
2.3. Flow Cytometry Analysis
2.4. RNA-Seq Analysis
2.5. In Silico Analysis of TF and TFBS
2.6. Construction of NKX3-1 Overexpressing Cell Lines
2.7. Quantitative Reverse Transcription PCR (RT-qPCR)
2.8. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Dumont, J.; Euwart, D.; Mei, B.; Estes, S.; Kshirsagar, R. Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Crit. Rev. Biotechnol. 2016, 36, 1110–1122. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.L.; Goh, J.B.; Srinivasan, H.; Liu, K.I.; Gowher, A.; Shanmugam, R.; Lim, H.L.; Choo, M.; Tang, W.Q.; Tan, A.H.; et al. A human expression system based on HEK293 for the stable production of recombinant erythropoietin. Sci. Rep. 2019, 9, 16768. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Baek, M.; Lee, G.M. Comprehensive characterization of dihydrofolate reductase-mediated gene amplification for the establishment of recombinant human embryonic kidney 293 cells producing monoclonal antibodies. Biotechnol. J. 2021, 16, e2000351. [Google Scholar] [CrossRef]
- Yoon, S.K.; Song, J.Y.; Lee, G.M. Effect of low culture temperature on specific productivity, transcription level, and heterogeneity of erythropoietin in Chinese hamster ovary cells. Biotechnol. Bioeng. 2003, 82, 289–298. [Google Scholar] [CrossRef]
- Fox, S.R.; Patel, U.A.; Yap, M.G.; Wang, D.I. Maximizing interferon-gamma production by Chinese hamster ovary cells through temperature shift optimization: Experimental and modeling. Biotechnol. Bioeng. 2004, 85, 177–184. [Google Scholar] [CrossRef]
- Lin, C.Y.; Huang, Z.; Wen, W.; Wu, A.; Wang, C.; Niu, L. Enhancing Protein Expression in HEK-293 Cells by Lowering Culture Temperature. PLoS ONE 2015, 10, e0123562. [Google Scholar] [CrossRef]
- Yoon, S.K.; Hwang, S.O.; Lee, G.M. Enhancing effect of low culture temperature on specific antibody productivity of recombinant Chinese hamster ovary cells: Clonal variation. Biotechnol. Prog. 2004, 20, 1683–1688. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, J.H.; Ha, T.K.; Samoudi, M.; Lewis, N.E.; Palsson, B.O.; Kildegaard, H.F.; Lee, G.M. Revealing Key Determinants of Clonal Variation in Transgene Expression in Recombinant CHO Cells Using Targeted Genome Editing. ACS Synth. Biol. 2018, 7, 2867–2878. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kildegaard, H.F.; Lewis, N.E.; Lee, G.M. Mitigating Clonal Variation in Recombinant Mammalian Cell Lines. Trends Biotechnol. 2019, 37, 931–942. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Novak, N.; Baumann, M.; Koehn, J.; Borth, N. Bioinformatic Identification of Chinese Hamster Ovary (CHO) Cold-Shock Genes and Biological Evidence of their Cold-Inducible Promoters. Biotechnol. J. 2020, 15, e1900359. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.; Akhtar, S.; McKenzie, E.A.; Dickson, A.J. Temperature Down-Shift Modifies Expression of UPR-/ERAD-Related Genes and Enhances Production of a Chimeric Fusion Protein in CHO Cells. Biotechnol. J. 2021, 16, e2000081. [Google Scholar] [CrossRef] [Green Version]
- Bedoya-López, A.; Estrada, K.; Sanchez-Flores, A.; Ramírez, O.T.; Altamirano, C.; Segovia, L.; Miranda-Ríos, J.; Trujillo-Roldán, M.A.; Valdez-Cruz, N.A. Effect of Temperature Downshift on the Transcriptomic Responses of Chinese Hamster Ovary Cells Using Recombinant Human Tissue Plasminogen Activator Production Culture. PLoS ONE 2016, 11, e0151529. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Kim, S.H.; Shin, S.W.; Grav, L.M.; Pedersen, L.E.; Lee, J.S.; Lee, G.M. Comprehensive Analysis of Genomic Safe Harbors as Target Sites for Stable Expression of the Heterologous Gene in HEK293 Cells. ACS Synth. Biol. 2020, 9, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [Green Version]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Kwak, J.M.; Lee, J.S. Endogenous p21-Dependent Transgene Control for CHO Cell Engineering. ACS Synth. Biol. 2020, 9, 1572–1580. [Google Scholar] [CrossRef]
- He, W.W.; Sciavolino, P.J.; Wing, J.; Augustus, M.; Hudson, P.; Meissner, P.S.; Curtis, R.T.; Shell, B.K.; Bostwick, D.G.; Tindall, D.J.; et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 1997, 43, 69–77. [Google Scholar] [CrossRef]
- Marchant, R.J.; Al-Fageeh, M.B.; Underhill, M.F.; Racher, A.J.; Smales, C.M. Metabolic rates, growth phase, and mRNA levels influence cell-specific antibody production levels from in vitro-cultured mammalian cells at sub-physiological temperatures. Mol. Biotechnol. 2008, 39, 69–77. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, M.H.; Min, H.; Lee, J.S. Enhancement of Transgene Expression by Mild Hypothermia Is Promoter Dependent in HEK293 Cells. Life 2021, 11, 901. https://doi.org/10.3390/life11090901
Jang MH, Min H, Lee JS. Enhancement of Transgene Expression by Mild Hypothermia Is Promoter Dependent in HEK293 Cells. Life. 2021; 11(9):901. https://doi.org/10.3390/life11090901
Chicago/Turabian StyleJang, Min Ho, Honggi Min, and Jae Seong Lee. 2021. "Enhancement of Transgene Expression by Mild Hypothermia Is Promoter Dependent in HEK293 Cells" Life 11, no. 9: 901. https://doi.org/10.3390/life11090901
APA StyleJang, M. H., Min, H., & Lee, J. S. (2021). Enhancement of Transgene Expression by Mild Hypothermia Is Promoter Dependent in HEK293 Cells. Life, 11(9), 901. https://doi.org/10.3390/life11090901






