Separation, Purification, Structure Analysis, In Vitro Antioxidant Activity and circRNA-miRNA-mRNA Regulatory Network on PRV-Infected RAW264.7 Cells of a Polysaccharide Derived from Arthrospira platensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Comparison of Different Extraction Methods
2.2.1. Different Extraction Methods of PAP
2.2.1.1. Hot Water Extraction Method
2.2.1.2. Enzyme-Assisted Hot Water Extraction Method
2.2.1.3. Ultrasonication-Assisted Hot Water Extraction Method
2.2.1.4. Microwave-Assisted Hot Water Extraction Method
2.2.1.5. Freeze-Thaw Assisted Hot Water Extraction Method
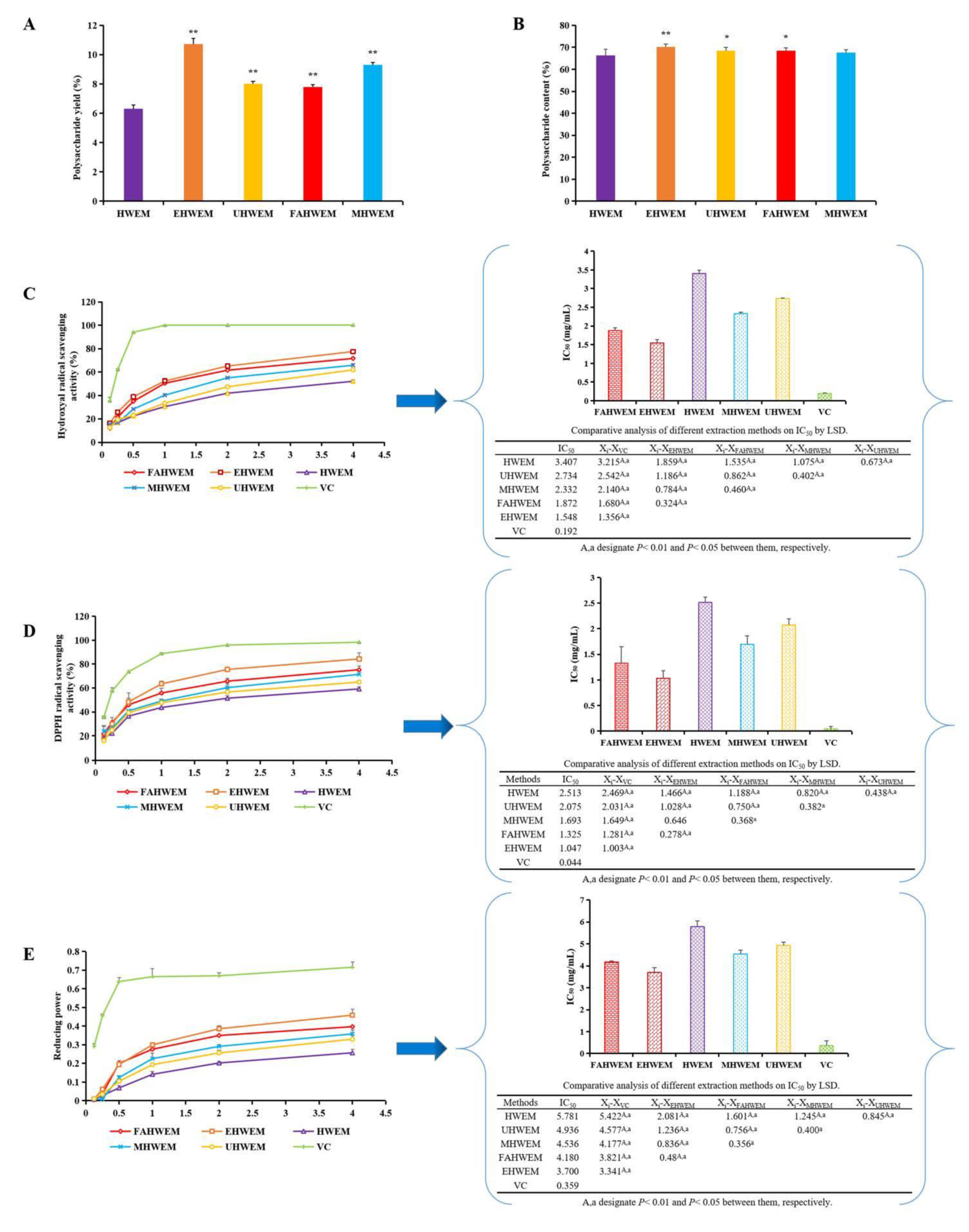
2.2.2. Comparison of Extraction Rate and Polysaccharide Content
2.2.3. Comparison of Antioxidant Activity
2.2.3.1. Determination of Hydroxyl Radical Scavenging Activity
2.2.3.2. Determination of DPPH Free Radical Scavenging Activity
2.2.3.3. Reducing Power
2.3. Separation and Purification of PAP
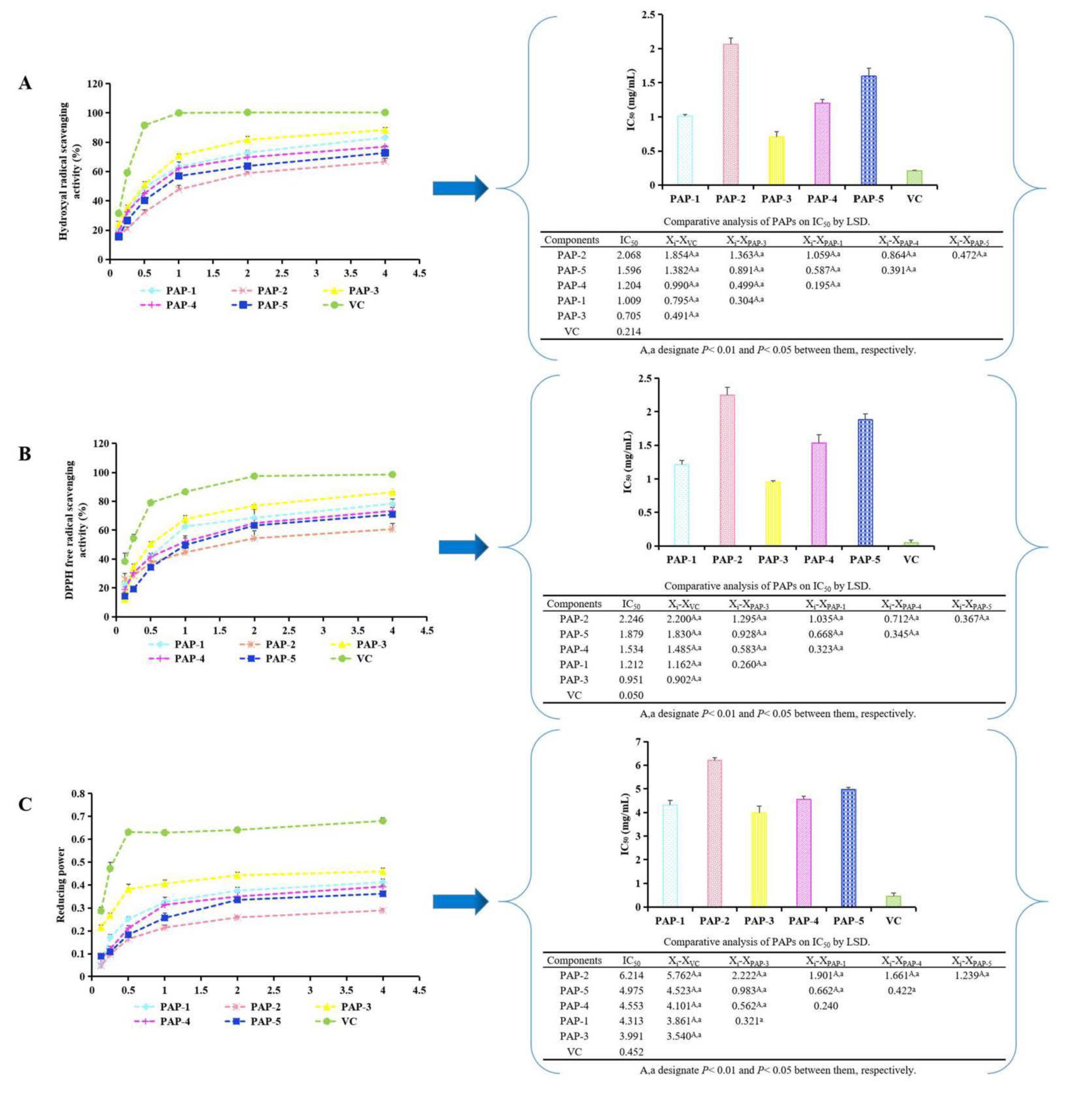
2.4. Comparison of Antioxidant Activity of PAP
2.5. Structure Analysis of the First Component of PAP (PAP-1)
2.5.1. Determination of Molecular Weight
2.5.2. Specific Rotation
2.5.3. FT-IR
2.5.4. Monosaccharide Composition Analysis
2.5.5. Periodate Oxidation and Smith Degradation
2.5.6. Methylation Analysis
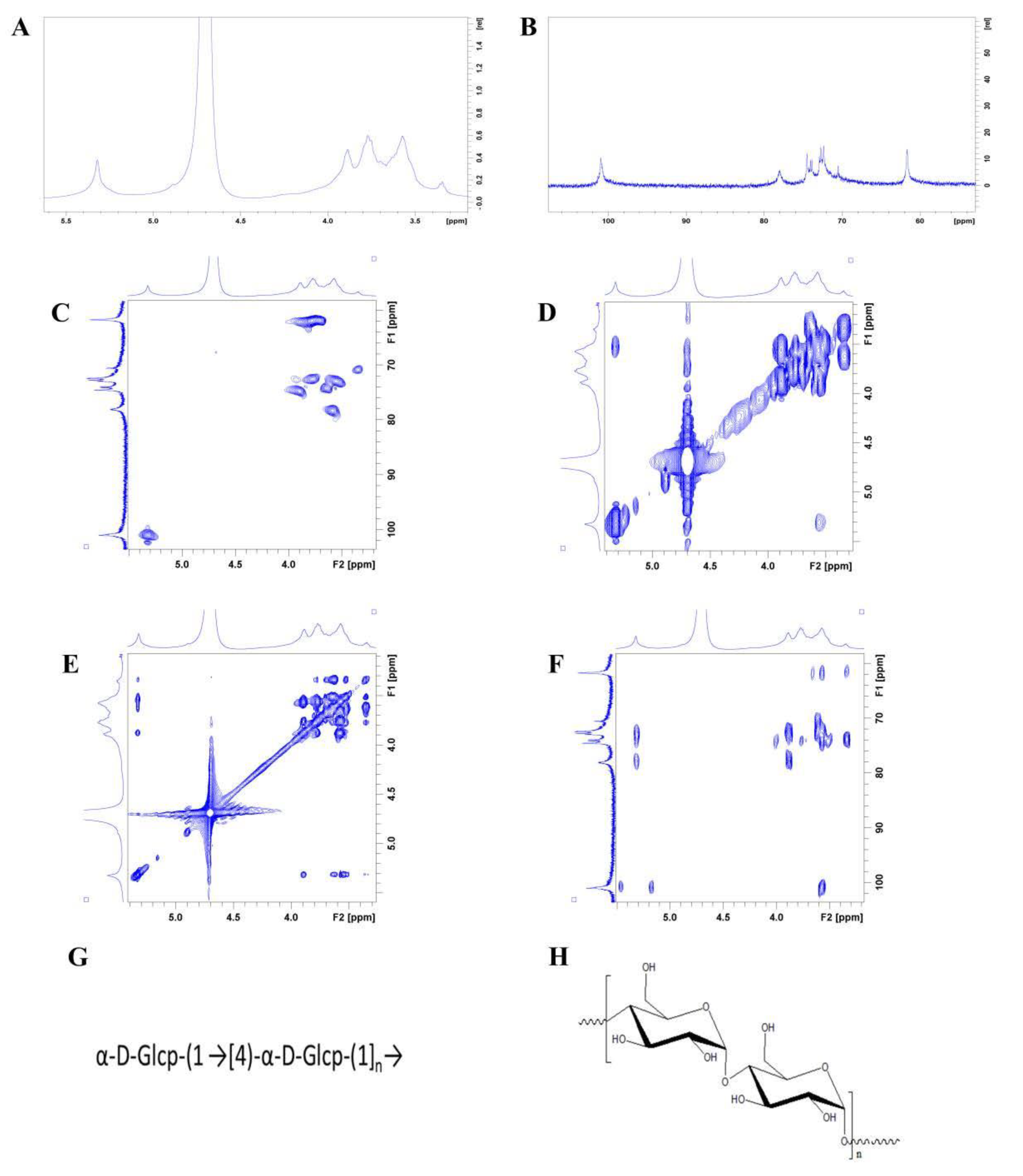
2.5.7. NMR Analysis
2.6. The Effect of PAP-1 on Antioxidation of PRV-Infected RAW264.7 Cells
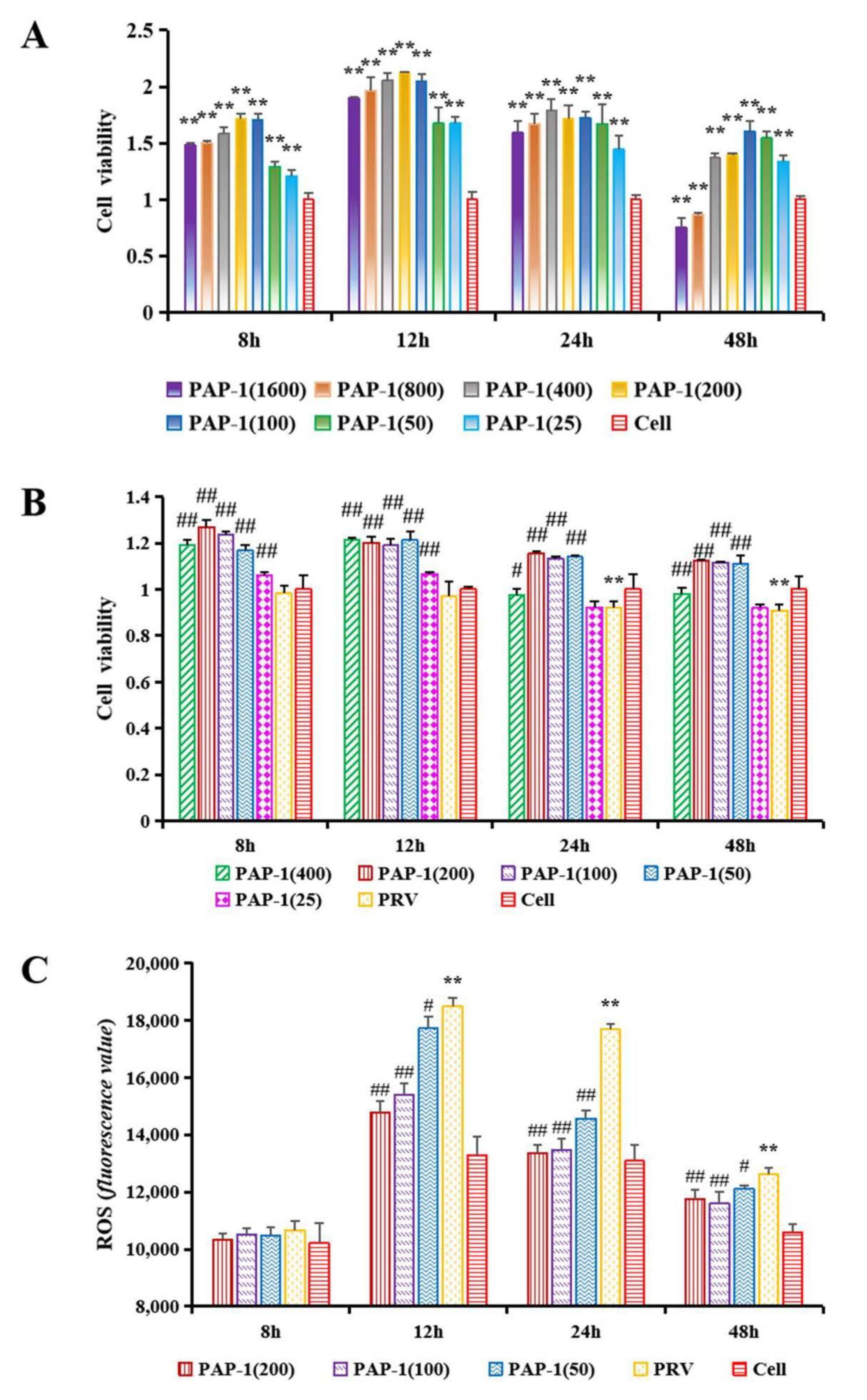
2.6.1. The Effect of PAP-1 on the Activity of RAW264.7 Cells
2.6.2. The Effect of PAP-1 on the Activity of PRV-Infected RAW264.7 Cells
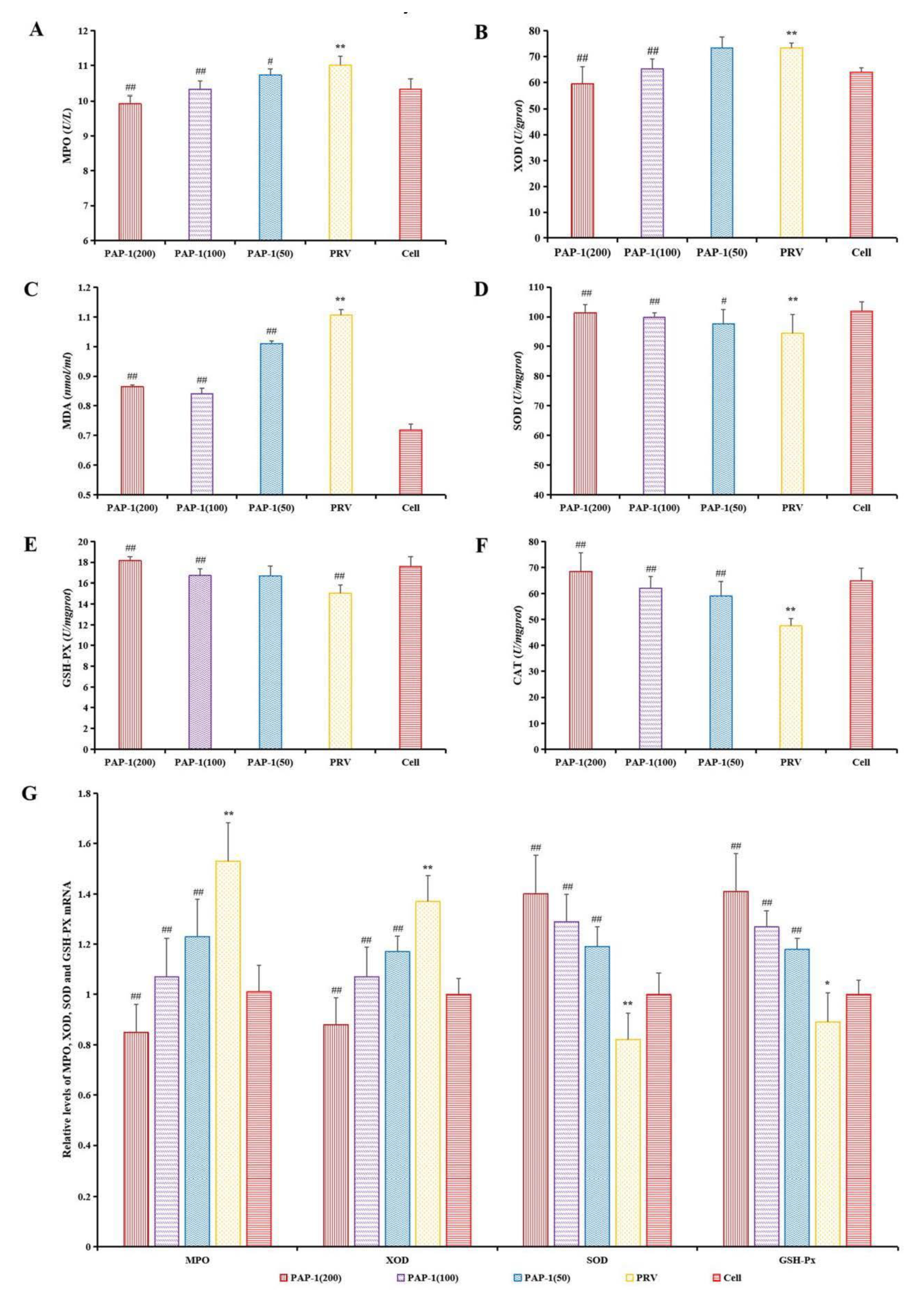
2.6.3. The Effect of PAP-1 on the Antioxidative Capacity in PRV-Infected RAW264.7 Cells
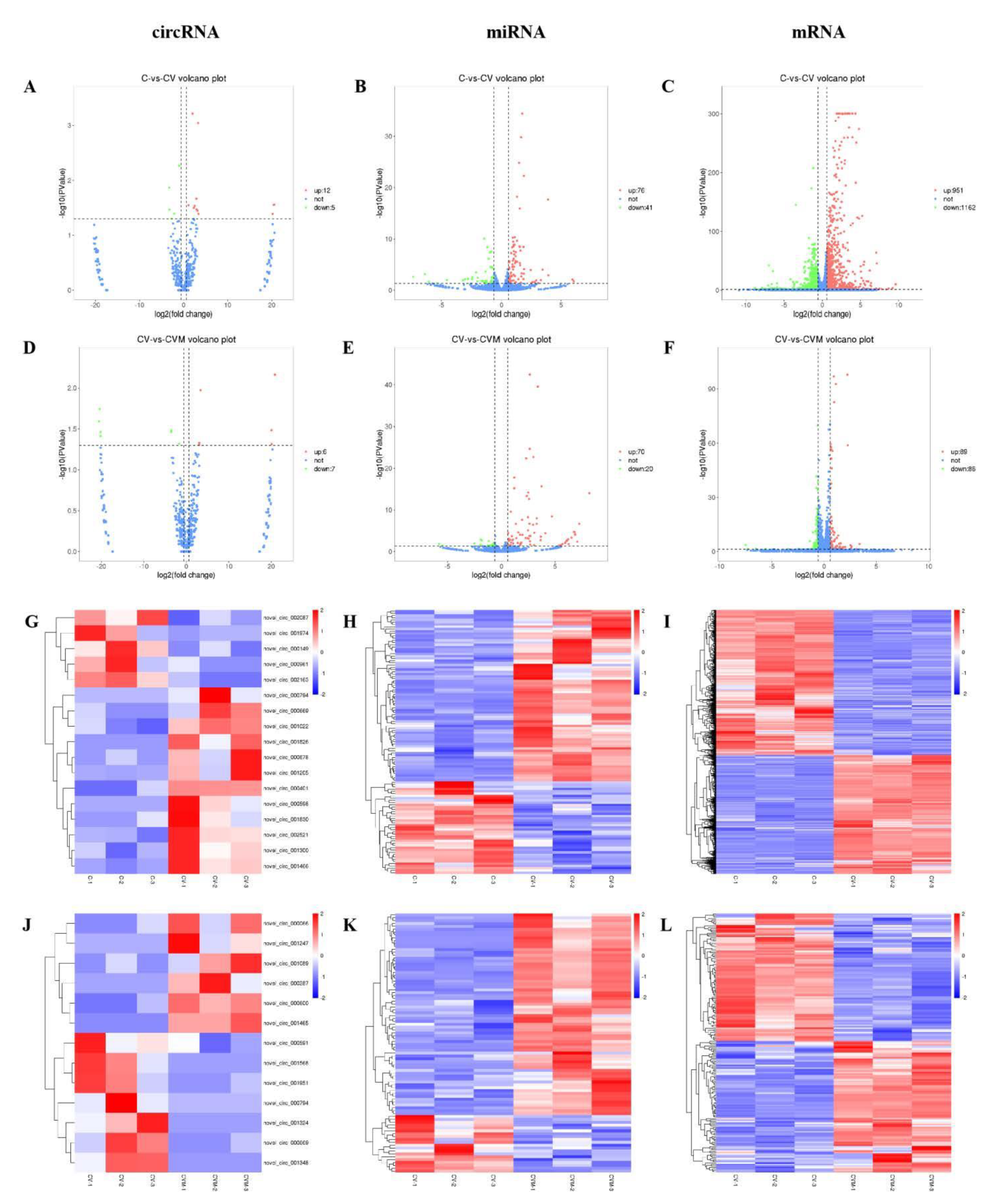
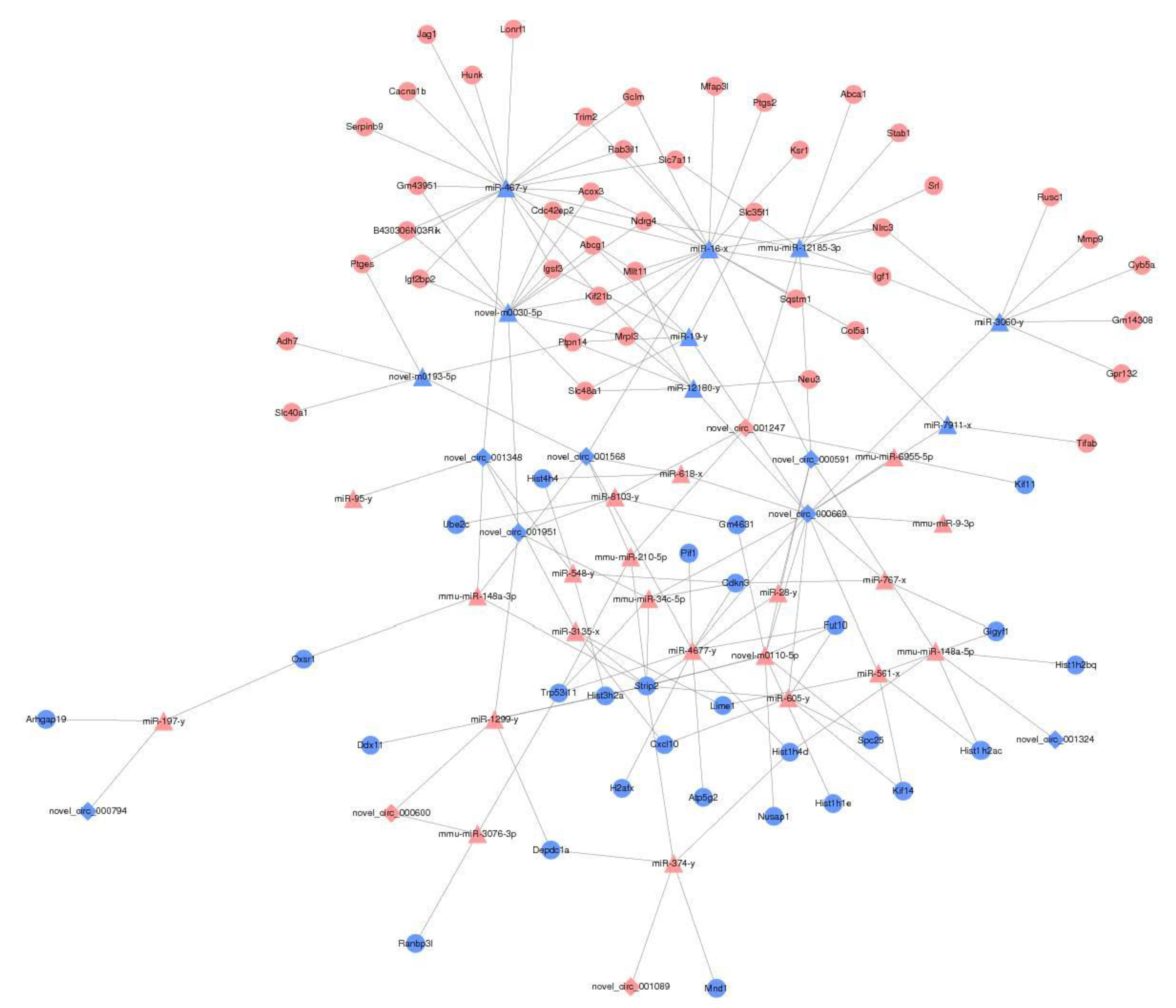
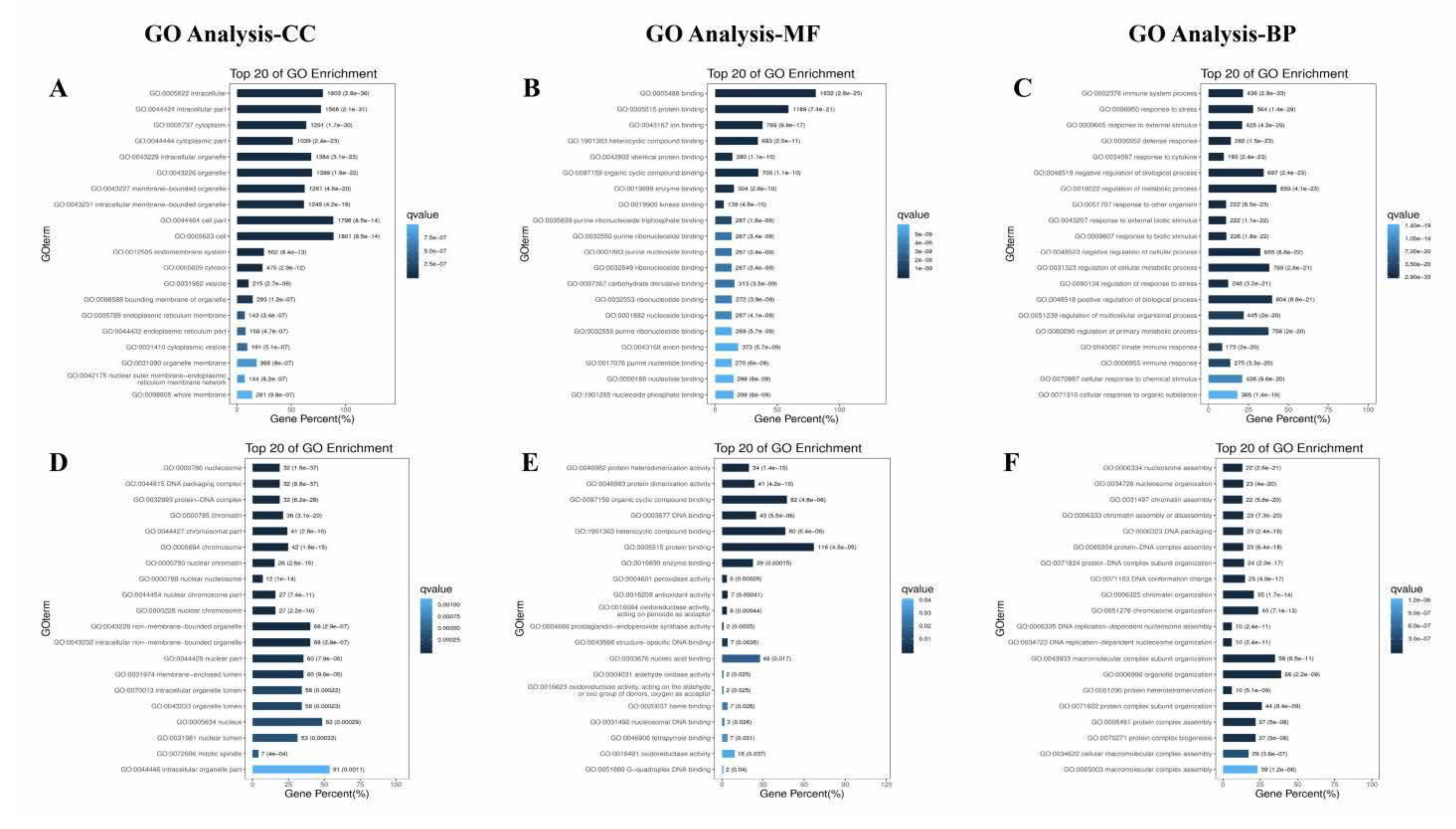
2.6.4. CeRNA-Seq
2.6.5. Quantitative Real-Time PCR
2.7. Statistical Analysis
3. Results and Discussion
3.1. Comparison of Different Extraction Methods
3.2. Isolation and Purification
3.3. Determination of the Antioxidant Activity of PAPs
3.4. Structure Analysis of PAP-1
3.4.1. Molecular Weight
3.4.2. Specific Optical Rotation
3.4.3. FT-IR
3.4.4. Analysis of Monosaccharide Composition
3.4.5. Periodate Oxidation and Smith Degradation
3.4.6. Methylation Analysis
3.4.7. NMR
3.5. The Effect of PAP-1 on the Activity of RAW264.7 Cells
3.6. The Effect of PAP-1 on the Activity of PRV-Infected RAW264.7 Cells
3.7. The Effect of PAP-1 on Antioxidant Capacity in PRV-Infected RAW264.7 Cells
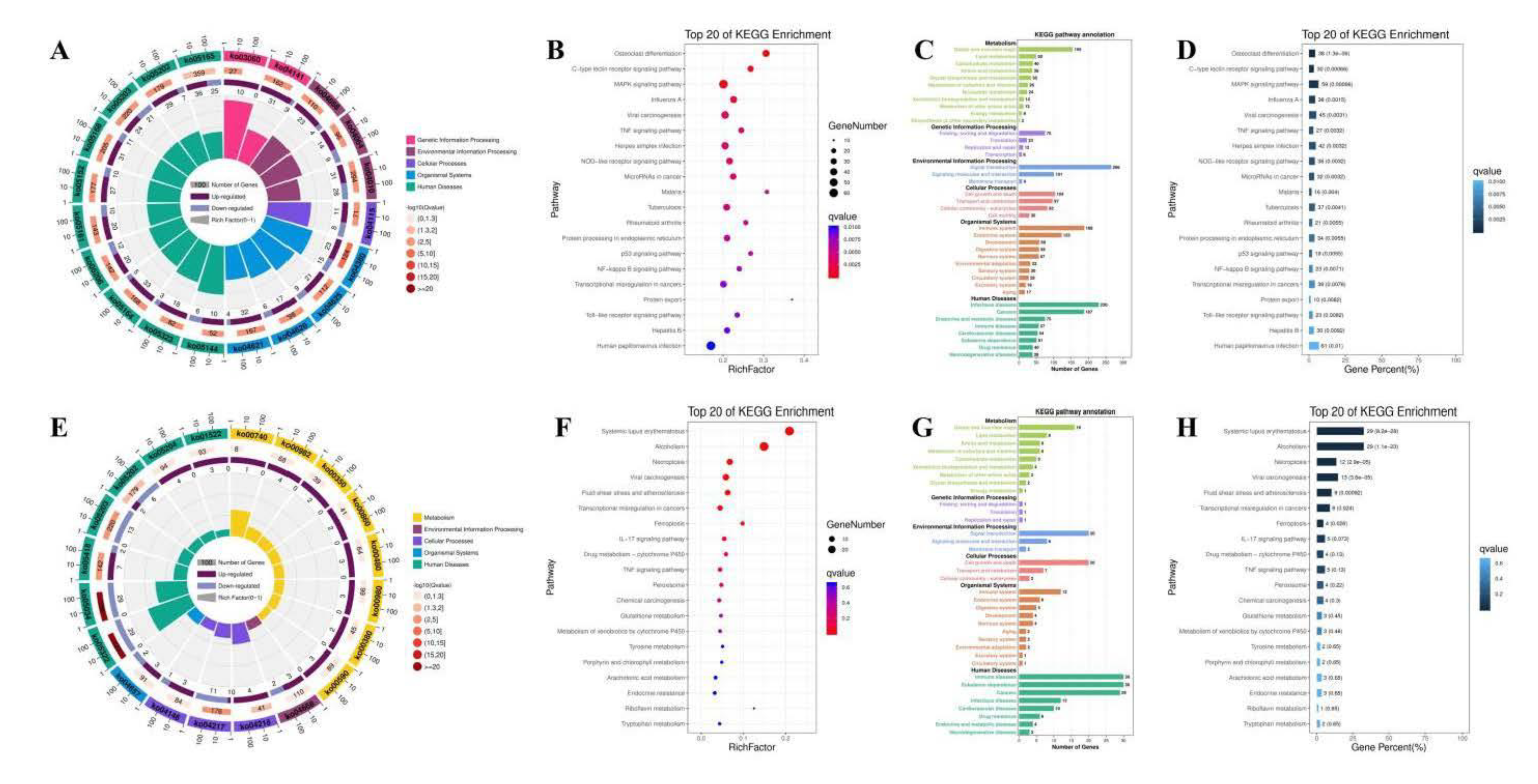
3.8. The Effect of PAP-1 on the Network of CircRNA-miRNA-mRNA in PRV-Infected RAW264.7 Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, B.Y.; Liu, Q.; Huang, Y.H.; Yuan, Y.L.; Ma, Q.Q.; Du, M.L.; Cai, T.G.; Cai, Y. Extraction of polysaccharide from Spirulina and evaluation of its activities. Evid.-Based. Compl. Alt. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Yan, X.T.; Liang, J.; Li, S.J.; He, H.R.; Xiong, Q.P.; Lai, X.P.; Hou, S.Z.; Huang, S. Comparison of different extraction methods for polysaccharides from Dendrobium officinale stem. Carbohydr. Polym. 2018, 198, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Chen, G.J.; Wang, Z.R.; Kan, J.Q. A comparison of a polysaccharide extracted from ginger (Zingiber officinale) stems and leaves using different methods: Preparation, structure characteristics, and biological activities. Int. J. Biol. Macromol. 2020, 151, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Desai, K.; Sivakami, S. Spirulina: The wonder Food of the 21st century. Asia. Pac. Biotech. News. 2004, 8, 1298–1302. [Google Scholar] [CrossRef]
- Chen, Y.H.; Wan, X.Z.; Wu, D.S.; Ouyang, Y.Z.; Gao, L.Y.; Chen, Z.X.; El-Seedi, H.R.; Wang, M.-F.; Chen, X.-H.; Zhao, C. Characterization of the structure and analysis of the anti-oxidant effect of microalga Spirulina platensis polysaccharide on Caenorhabditis elegans mediated by modulating microRNAs and gut microbiota. Int. J. Biol. Macromol. 2020, 163, 2295–2305. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.T.; Xiong, H.Y.; Zhu, X.L.; Ji, C.L.; Xue, J.N.; Li, R.Z.; Ge, B.S.; Cui, H.L. Polysaccharide from Spirulina platensis ameliorates diphenoxylate-induced constipation symptoms in mice. Int. J. Biol. Macromol. 2019, 133, 1090–1101. [Google Scholar] [CrossRef]
- Zhang, F.; Lu, J.; Zhang, J.G.; Xie, J.X. Protective effects of a polysaccharide from Spirulina platensis on dopaminergic neurons in an MPTP-induced Parkinson’s disease model in C57BL/6J mice. Neural. Regen. Res. 2015, 10, 308–313. [Google Scholar] [CrossRef]
- Mader, J.; Gallo, A.; Schommartz, T.; Handke, W.; Nagel, C.H.; Günther, P.; Brune, W.; Reich, K. Calcium spirulan derived from Spirulina platensis inhibits herpes simplex virus 1 attachment to human keratinocytes and protects against herpes labialis. J. Allergy. Clin. Immun. 2016, 137, 197–203.e3. [Google Scholar] [CrossRef] [Green Version]
- Rajasekar, P.; Palanisamy, S.; Anjali, R.; Vinosha, M.; Elakkiya, M.; Marudhupandi, T.; Tabarsa, M.; You, S.G.; Prabhu, N.M. Isolation and structural characterization of sulfated polysaccharide from Spirulina platensis and its bioactive potential: In vitro antioxidant, antibacterial activity and Zebrafish growth and reproductive performance. Int. J. Biol. Macromol. 2019, 141, 809–821. [Google Scholar] [CrossRef]
- Wu, X.Y.; Liu, Z.C.; Liu, Y.; Yang, Y.; Shi, F.L.; Cheong, K.L.; Teng, B. Immunostimulatory effects of polysaccharides from Spirulina platensis in vivo and vitro and their activation mechanism on RAW 246.7 macrophages. Mar. Drugs 2020, 18, 538. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.W.; Yin, J.C.; Ren, X.F. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antivir. Res. 2010, 85, 346–353. [Google Scholar] [CrossRef]
- Luo, P.; Liu, D.; Li, J. Pharmacological perspective: Glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105995. [Google Scholar] [CrossRef]
- Liu, H.; You, L.M.; Wu, J.; Zhao, M.F.; Guo, R.; Zhang, H.L.; Su, R.; Mao, Q.; Deng, D.; Hao, Y. Berberine suppresses influenza virus-triggered NLRP3 inflammasome activation in macrophages by inducing mitophagy and decreasing mitochondrial ROS. J. Leukoc. Biol. 2020, 108, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.J.; Jassey, A.; Liu, C.H.; Tai, C.J.; Richardson, C.D.; Wong, S.H.; Lin, L.T. Targeting autophagy augments bbr-mediated cell death in human hepatoma cells harboring hepatitis c virus RNA. Cells 2020, 9, 908. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Shen, M.Y.; Song, Q.Q.; Xie, J.H. Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review. Carbohydr. Polym. 2018, 183, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Lammi, C.; Arnoldi, A. Food-derived antioxidants and COVID-19. J. Food. Biochem. 2020, 45, e13557. [Google Scholar] [CrossRef]
- Li, L.Y.; Huang, T.; Lan, C.; Ding, H.; Yan, C.S.; Dou, Y.L. Protective effect of polysaccharide from Sophora japonica L. flower buds against UVB radiation in a human keratinocyte cell line (HaCaT cells). J. Photoch. Photobio. B. 2018, 191, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Han, R.H.; Tang, F.T.; Lu, M.L.; Xu, C.H.; Hu, J.; Mei, M.; Wang, H.X. Astragalus polysaccharide ameliorates H2O2-induced human umbilical vein endothelial cell injury. Mol. Med. Rep. 2017, 15, 4027–4034. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hu, T.J.; Liu, H.L.; Shuai, X.H. Inhibitory effect of sargassum polysaccharide on oxidative stress induced by infectious bursa disease virus in chicken bursal lymphocytes. Int. J. Biol. Macromol. 2011, 49, 607–615. [Google Scholar] [CrossRef]
- Hirahashi, T.; Matsumoto, M.; Hazeki, K.; Saeki, Y.; Ui, M.; Seya, T. Activation of the human innate immune system by Spirulina: Augmentation of interferon production and NK cytotoxicity by oral administration of hot water extract of Spirulina platensis. Int. Immunopharmacol. 2002, 2, 423–434. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Bhattacharyya, S.N.; Habermacher, R.; Martine, U.; Closs, E.I.; Filipowicz, W. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006, 125, 1111–1124. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.W.; Tsai, K.N.; Chen, Y.S.; Chang, R.Y. Differential miRNA Expression Profiling Reveals Correlation of miR125b-5p with Persistent Infection of Japanese Encephalitis Virus. Int. J. Mol. Sci. 2021, 22, 4218. [Google Scholar] [CrossRef]
- Rahman, A.; Susmi, T.F.; Yasmin, F.; Bhattacharjee, A.; Hossain, M.U.; Das, K.C.; Keya, C.A.; Salimullah, M. Unveiling ancestral relations, host–pathogen interactions and comparative viral miRNA insights of dengue virus serotypes. Netw. Model. Anal. Health 2021, 10, 1–13. [Google Scholar]
- Zhang, X.Q.; Li, C.; Zhang, B.Z.; Li, Z.H.; Zeng, W.; Luo, R.; Cao, J.Y.; Cheng, G.F.; Fan, S.X.; He, Q.G. Differential expression and correlation analysis of miRNA–mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci. Rep. 2020, 11, 1868. [Google Scholar] [CrossRef]
- Zhong, Y.X.; Du, Y.J.; Yang, X.; Mo, Y.Z.; Fan, C.; Xiong, F.; Ren, D.X.; Ye, X.; Li, C.W.; Wang, Y.M.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Lin, F.; Chen, H.W.; Zhao, G.A.; Li, Y.; He, X.H.; Liang, W.Q.; Shi, Z.L.; Sun, S.Y.; Tian, P.P.; Huang, M.Y.; et al. Advances in Research on the circRNA-miRNA-mRNA Network in Coronary Heart Disease Treated with Traditional Chinese Medicine. Evid.-Based Complement. Altern. Med. 2020, 2020, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Sokolova, E.V.; Barabanova, A.O.; Bogdanovich, R.N.; Khomenko, V.A.; Solov’eva, T.F.; Yermak, I.M. In vitro antioxidant properties of red algal polysaccharides. Biomed. Prev. Nut. 2011, 1, 161–167. [Google Scholar] [CrossRef]
- Bai, R.G.; Tuvikene, R. Potential antiviral properties of industrially important marine algal polysaccharides and their significance in fighting a future viral pandemic. Viruses 2021, 13, 1817. [Google Scholar]
- Pujol, C.A.; Ray, S.; Ray, B.; Damonte, E.B. Antiviral activity against dengue virus of diverse classes of algal sulfated polysaccharides. Int. J. Biol. Macromol. 2012, 51, 412–416. [Google Scholar] [CrossRef]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Li, B.; Zhang, N.; Wang, D.X.; Jiao, L.L.; Tan, Y.; Wang, J.; Li, H.; Wu, W.; Jiang, D.C. Structural analysis and antioxidant activities of neutral polysaccharide isolated from Epimedium koreanum Nakai. Carbohydr. Polym. 2018, 196, 246–253. [Google Scholar] [CrossRef]
- Ye, Z.P.; Wang, W.; Yuan, Q.X.; Ye, H.; Sun, Y.; Zhang, H.C.; Zeng, X.X. Box-Behnken design for extraction optimization, characterization and in vitro antioxidant activity of Cicer arietinum L. hull polysaccharides. Carbohydr. Polym. 2016, 147, 354–364. [Google Scholar] [CrossRef]
- Sahragard, N.; Jahanbin, K. Structural elucidation of the main water-soluble polysaccharide from Rubus anatolicus roots. Carbohydr. Polym. 2017, 175, 610–617. [Google Scholar] [CrossRef]
- Huang, H.Y.; Liu, J.J.; Xi, R.R.; Xing, X.M.; Yuan, J.H.; Yang, L.Q.; Tao, G.H.; Gong, C.M.; Zhuang, Z.X. An investigation of hormesis of trichloroethylene in L-02 liver cells by differential proteomic analysis. Mol. Biol. Rep. 2009, 36, 2119–2129. [Google Scholar] [CrossRef]
- Ju, T.; Xi, J. Continuous extraction optimization, molecular structures and antioxidant activities of polysaccharide from gracilariopsis lemaneiformis using liquid-phase pulsed discharge. Sep. Purif. Technol. 2020, 236, 116241. [Google Scholar] [CrossRef]
- Chen, S.H.; Huang, H.L.; Huang, G.L. Extraction, derivatization and antioxidant activity of cucumber polysaccharide. Int. J. Biol. Macromol. 2019, 140, 1047–1053. [Google Scholar] [CrossRef]
- Dammak, M.I.; Salem, Y.B.; Belaid, A.; Mansour, H.B.; Hammami, S.; Cerf, D.L.; Majdoub, H. Partial characterization and antitumor activity of a polysaccharide isolated from watermelon rinds. Int. J. Biol. Macromol. 2019, 136, 632–641. [Google Scholar] [CrossRef]
- Ren, Y.P.; Liu, S.X. Effects of separation and purification on structural characteristics of polysaccharide from quinoa (chenopodium quinoa willd). Biochem. Biophys. Res. Commun. 2020, 522, 286–291. [Google Scholar] [CrossRef]
- Jahanbin, K.; Abbasian, A.; Ahang, M. Isolation, purifification and structural characterization of a new water-soluble polysaccharide from Eremurus stenophyllus (boiss. & buhse) baker roots. Carbohydr. Polym. 2017, 178, 386–393. [Google Scholar] [PubMed]
- Tang, Y.; Zhu, Z.Y.; Liu, Y.; Sun, H.Q.; Song, Q.Y.; Zhang, Y.M. The chemical structure and anti-aging bioactivity of an acid polysaccharide obtained from rose buds. Food. Funct. 2018, 9, 2300–2312. [Google Scholar] [CrossRef]
- Zhang, J.J.; Meng, G.Y.; Zhai, G.Y.; Yang, Y.H.; Zhao, H.J.; Jia, L. Extraction, characterization and antioxidant activity of polysaccharides of spent mushroom compost of Ganoderma lucidum. Int. J. Biol. Macromol. 2016, 82, 432–439. [Google Scholar] [CrossRef]
- Zhu, Z.Y.; Liu, N.; Si, C.L.; Liu, Y.; Ding, L.N.; Jing, C.; Liu, A.J.; Zhang, Y.M. Structure and anti-tumor activity of a high-molecular-weight polysaccharide from cultured mycelium of Cordyceps gunnii. Carbohydr. Polym. 2012, 88, 1072–1076. [Google Scholar] [CrossRef]
- Chaiklahan, R.; Chirasuwan, N.; Triratana, P.; Loha, V.; Tia, S.; Bunnag, B. Polysaccharide extraction from Spirulina sp. and its antioxidant capacity. Int. J. Biol. Macromol. 2013, 58, 73–78. [Google Scholar] [CrossRef]
- Steinhorn, G.; Sims, I.M.; Carnachan, S.M.; Carr, A.J.; Schlothauer, R. Isolation and characterisation of arabinogalactan-proteins from new zealand kanuka honey. Food. Chem. 2011, 128, 949–956. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.Q.; Yang, S.; Lu, Z.H.; Li, G.L.; Liu, J.W.; Zhou, B.; Wu, D.R.; Wang, L. Isolation, Purification, Characterization, and Immunomodulatory Activity Analysis of α-Glucans from Spirulina platensis. ACS. Omega 2021, 6, 21384–21394. [Google Scholar] [CrossRef]
- Nie, C.Z.P.; Zhu, P.L.; Ma, S.P.; Wang, M.C.; Hu, Y.D. Purification, characterization and immunomodulatory activity of polysaccharides from stem lettuce. Carbohydr. Polym. 2018, 188, 236–242. [Google Scholar] [CrossRef]
- Sun, Z.L.; Peng, Y.; Zhao, W.W.; Xiao, L.L.; Yang, P.M. Purification, characterization and immunomodulatory activity of a polysaccharide from Celosia cristata. Carbohydr. Polym. 2015, 133, 337–344. [Google Scholar] [CrossRef]
- Wang, M.L.; Hou, Y.Y.; Chiu, Y.S.; Chen, Y.H. Immunomodulatory activities of Gelidium amansii gel extracts on murine RAW 264.7 macrophages. J. Food. Drug. Anal. 2013, 21, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.P.; Zi, W.; Pei, Z.F.; Liu, S. Characterization of polysaccharides and their antioxidant properties from Plumula nelumbinis. Saudi. Pharm. J. 2018, 26, 656–664. [Google Scholar] [CrossRef]
- Antunes, M.A.; Lopes-Pacheco, M.; Rocco, P.R.M. Oxidative stress-derived mitochondrial dysfunction in chronic obstructive pulmonary disease: A concise review. Oxid. Med. Cell. Longev. 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I.; Mano, J. Lipid peroxidation-derived reactive carbonyl species (RCS): Their interaction with ROS and cellular redox during environmental stresses. Environ. Exp. Bot. 2019, 165, 139–149. [Google Scholar] [CrossRef]
- Chen, S.; Chen, H.S.; Du, Q.H.; Shen, J.G. Targeting myeloperoxidase (MPO) mediated oxidative stress and inflammation for reducing brain ischemia injury: Potential application of natural compounds. Front. Physiol. 2020, 11, 433. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, G.W.; Liao, Y.J.; Gong, D.M. Myricetin inhibits the generation of superoxide anion by reduced form of xanthine oxidase. Food. Chem. 2017, 221, 1569–1577. [Google Scholar] [CrossRef]
- Fernandes, I.G.; Brito, C.A.D.; Reis, V.M.S.D.; Sato, M.N.; PereiRa, N.Z. SARS-CoV-2 and other respiratory viruses: What does oxidative stress have to do with it? Oxid. Med. Cell. Longev. 2020, 2020, 1–13. [Google Scholar] [CrossRef]
- Zeraik, M.L.; Serteyn, D.; Deby-Dupont, G.; Wauters, J.N.; Tits, M.; Yariwake, J.H.; Angenot, L.; Franck, T. Evaluation of the antioxidant activity of passion fruit (Passiflora edulis and Passiflora alata) extracts on stimulated neutrophils and myeloperoxidase activity assays. Food. Chem. 2011, 128, 259–265. [Google Scholar] [CrossRef]
- Liao, Z.Z.; Zhang, J.Y.; Liu, B.; Yan, T.X.; Xu, F.X.; Xiao, F.; Wu, B.; Bi, K.S.; Jia, Y. Polysaccharide from okra (abelmoschus esculentus (l.) moench) improves antioxidant capacity via pi3k/akt pathways and nrf2 translocation in a type 2 diabetes model. Molecules. 2019, 24, 1906. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.L.; Chen, N.; Cai, H.; Tang, Y.; Zhou, X.Y.; Huang, Y.; Gong, M.X.; Qin, C.Y.; Wei, X.W.; Qi, S.H. Analysis of momordica charantia polysaccharide components and their effects on ka-induced oxidative stress and neuronal loss in the hippocampus of epileptic rats. World. J. Neurosci. 2018, 8, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Pang, F.; Zhang, M.M.; Yang, X.J.; Li, G.H.; Zhu, S.; Nie, X.; Cao, R.Y.; Yang, X.H.; Zhang, Z.X.; Huang, H.F.; et al. Genome-wide analysis of circular RNAs in goat skin fibroblast cells in response to Orf virus infection. PeerJ. 2019, 7, e6267. [Google Scholar] [CrossRef]
- Zhao, W.; Su, J.Y.; Wang, N.N.; Zhao, N.Y.; Su, S. Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus. Int. J. Mol. Sci. 2021, 22, 6537. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Qiu, L.L.; Bai, M.; Wang, L.D.; Hu, X.D.; Huang, L.; Chen, G.H.; Chang, G.B. Identification, biogenesis and function prediction of novel circRNA during the chicken ALV-J infection. Anim. Biotechnol. 2020, 10, 1–11. [Google Scholar]
- Zhang, H.L.; Chen, Z.M.; Zhong, Z.Y.; Gong, W.F.; Li, J. Total saponins from the leaves of Panax notoginseng inhibit depression on mouse chronic unpredictable mild stress model by regulating circRNA expression. Brain. Behav. 2018, 8, e01127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werfel, S.; Nothjunge, S.; Schwarzmayr, T.; Strom, T.M.; Meitinger, T.; Engelhardt, S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell Cardiol. 2016, 98, 103–107. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, M.-X.; Xie, X.-D.; Wang, X.-R.; Hu, W.-Y.; Zhao, Y.; Chen, Q.; Ji, L.; Wei, Y.-Y.; Yu, M.-L.; Hu, T.-J. Separation, Purification, Structure Analysis, In Vitro Antioxidant Activity and circRNA-miRNA-mRNA Regulatory Network on PRV-Infected RAW264.7 Cells of a Polysaccharide Derived from Arthrospira platensis. Antioxidants 2021, 10, 1689. https://doi.org/10.3390/antiox10111689
Cao M-X, Xie X-D, Wang X-R, Hu W-Y, Zhao Y, Chen Q, Ji L, Wei Y-Y, Yu M-L, Hu T-J. Separation, Purification, Structure Analysis, In Vitro Antioxidant Activity and circRNA-miRNA-mRNA Regulatory Network on PRV-Infected RAW264.7 Cells of a Polysaccharide Derived from Arthrospira platensis. Antioxidants. 2021; 10(11):1689. https://doi.org/10.3390/antiox10111689
Chicago/Turabian StyleCao, Mi-Xia, Xiao-Dong Xie, Xin-Rui Wang, Wen-Yue Hu, Yi Zhao, Qi Chen, Lu Ji, Ying-Yi Wei, Mei-Ling Yu, and Ting-Jun Hu. 2021. "Separation, Purification, Structure Analysis, In Vitro Antioxidant Activity and circRNA-miRNA-mRNA Regulatory Network on PRV-Infected RAW264.7 Cells of a Polysaccharide Derived from Arthrospira platensis" Antioxidants 10, no. 11: 1689. https://doi.org/10.3390/antiox10111689





