Abstract
Human dental pulp stem cells (hDPSCs), which are characterized by self-renewal capacity and the ability of multilineage differentiation, have gained increased attention in regenerative medicine recently. Histone acetylation modulator proteins (HAMPs) are a protein family that mediates the modification and identification of histone acetylation and participates in various critical cellular processes. Here, we comprehensively surveyed the expression profile of HAMPs during osteoblast differentiation of hDPSCs and found that the HDAC class III pathway was upregulated, whereas the signal transducer and activator of transcription 3 (STAT3) signaling was downregulated during osteogenesis. Further laboratory research demonstrated that Sirtuin-1 (SIRT1), a class III HDAC, was upregulated and STAT3 activation was downregulated during osteogenic differentiation. SIRT1 counteracted the activation of STAT3 to promote osteogenic differentiation of hDPSCs at 7 and 21 days in both Western blot assay and chemical staining, which highlights the promising utility of SIRT1 activators in hDPSCs-based therapies for bone augmentation strategies and provides clinical insights that may lead to the development of osteogenic agents.
1. Introduction
Human dental pulp stem cells (hDPSCs), currently considered to be a type of mesenchymal stem cells (MSCs), are isolated from dental pulp tissue, which has excellent self-renewal capacity and the ability of multidirectional differentiation [1,2,3]. These cells can differentiate into odontoblasts, chondrocytes, adipocytes, or osteoblasts under proper environmental conditions or certain induction and might be useful in oral and maxillofacial reconstruction, orofacial bone regeneration, and periodontal treatment [4,5]. hDPSCs have gained increased attention in tissue regeneration applications such as bone engineering, thanks to their multidirectional differentiation potency, easy availability, cost-effectiveness, cryopreserved ability, fewer side effects, and lack of ethical concerns [6,7]. The proliferation and osteogenic differentiation of hDPSCs need to be enhanced as much as possible to fulfill the demands of tissue engineering. Unfortunately, the exact mechanism of osteogenic differentiation is not fully understood, which might be a crucial barrier for the clinical implementation of hDPSCs.
The lineage commitment and differentiation process of hDPSCs are under both genetic and epigenetic control. Epigenetic regulation of DNA and DNA-related histone has been shown to regulate the phenotype, regeneration, and multidirectional differentiation of stem cells [8]. Histone acetylation modulator proteins (HAMPs) are a protein family that could modify and identify histone acetylation, which fall into classes including histone acetyltransferases (HATs, writers), histone deacetylases (HDACs, erasers), and readers [9,10,11,12]. A large and growing body of literature has recognized the importance of HAMPs in both the physiological and pathological processes, such as cellular differentiation, organism development, cancer, inflammation, and metabolic disease [10,13]. However, research has yet to systematically investigate the HAMPs during osteoblast differentiation of hDPSCs.
Sirtuin-1 (SIRT1) from class III HDAC is a kind of deacetylates protein exerting varieties of physiological functions containing DNA repair, cellular stress resistance, cell death threshold modulation, and metabolic parameters [14,15,16]. Although it is now well-established that SIRT1 regulates the transcription of signal transducer and activator of transcription 3 (STAT3), a transcription factor that participates in various critical cellular processes [17,18,19], the functional significance of SIRT1 during osteogenic differentiation of hDPSCs remains ill-defined, and the role of the SIRT1-STAT3 axis in osteogenic differentiation remains unclear.
In this report, we comprehensively surveyed the expression profile of HAMPs during osteoblast differentiation of hDPSCs. Our research demonstrated that SIRT1 counteracted the activation of STAT3 and promoted the osteogenic differentiation of hDPSCs at both early and late stages, with a train of bioinformatics analyses and laboratory findings. These findings emphasize the importance of the SIRT1 agonist in osteogenic differentiation, which may lead to the discovery of osteogenic agents.
2. Materials and Methods
2.1. hDPSCs’ Isolation and Culture
This research was approved and supervised by the Institutional Review Board of Nanjing Stomatological Hospital (KY-2020NL-029, Nanjing, China). Orthodontic impacted premolar teeth were collected from adults (aged 18–25 years) at Nanjing Stomatological Hospital. Five patients and ten premolars were included in the study. Prior to the study, each participant provided informed consent. Primary human pulp stem cells were isolated by both the explant and the enzymatic digestion method. Tooth surfaces were cleaned off and split along the long axis of the teeth by sterile instruments (NSK, Tokyo, Japan) under aseptic conditions. The pulp was extracted from the exposed pulp chamber using sterile tweezers and minced into tiny pieces (<1 mm3), and digested in 5 mg/mL collagenase type P (Roche, Basel, Switzerland) for 1 h at 37 °C, with vortex every 10 min to help tissue disruption. Cells were seeded in culture dishes with growth medium (α-MEM (Gibco, Grand Island, NY, USA), supplemented with 10% fetal calf serum (Hyclone, Logan, UT, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA)) in a moist atmosphere of 95% air and 5% CO2 at 37 °C after centrifugation. Cells were split when they reached 80% confluence. The fourth-passage cells were used in all experiments in this paper except for the FACS analysis.
2.2. Characterization of hDPSCs
hDPSCs were characterized by differentiation potential assessment and the surface marker test. hDPSCs were cultured in osteogenic media for osteogenic differentiation and chondrogenic differentiation according to the following protocol. hDPSCs were identified via their expression of cell surface antigens by flow cytometric analysis, including CD90, CD105, CD34, and CD45.
2.3. Osteogenic Differentiation
For osteogenic differentiation, adherent hDPSCs were seeded on 6-well plates at a density of 1 × 104 cells/mL and cultured in the osteogenic induction medium (α-MEM containing 20% fetal calf serum (Hyclone, Logan, UT, USA), 100 U/mL penicillin, 100 μg/mL streptomycin, 0.1 μM dexamethasone (Solarbio, Beijing, China), and 30 μg/mL L-Ascorbic Acid 2-phosphate (Sigma, Saint Louis, MO, USA)). The medium was replaced every 3 days during the incubation period.
2.4. Chondrogenic Differentiation
To induce chondrogenic differentiation, adherent hDPSCs were transferred to a 15 mL centrifuge tube containing chondrogenic induction medium (α-MEM containing 20% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 10−7 M dexamethasone, 10 ng/mL TGF β(c) (Sigma, Saint Louis, MO, USA), 50 µg/mL AA2P, and 1% ITS (Sigma, Saint Louis, MO, USA)). The medium was also changed every 3 days. After 14 days of culture, chondrogenic differentiation was confirmed by Alcian Blue staining (Solarbio, Beijing, China).
2.5. Alkaline Phosphatase (ALP) Staining and Von Kossa Staining
After 7 days of culture, differentiation of the cells into osteoblasts was confirmed by ALP staining (Solarbio, Beijing, China). After 21 days of culture, calcification in osteoblasts was detected by von Kossa staining (Solarbio, Beijing, China). hDPSCs were subjected to ALP staining or von Kossa staining using the corresponding assay kit, according to the standard directions, and photographed under a microscope.
2.6. Flow Cytometry (FCM) Analysis
The first-passage hDPSCs were harvested and washed twice with PBS and incubated with the following antibodies (all from BD Biosciences): CD90 (Clone 5E10, #561971), CD105 (Clone 266, #562408), CD34 (Clone 581, # 560942), and CD45 (Clone HI30, # 555482). The cells were labeled according to manufacturer’s protocols. Matched labeled isotypes were used as controls. We used a FACS Calibur instrument (BD Biosciences, Franklin Lakes, NJ, USA) to subject the samples and analyze the data with FlowJo software.
2.7. Western Blot
Proteins were retrieved from hDPSCs in RIPA lysis buffer (Beyotime), then subjected to SDS gel electrophoresis and electrophoretically transferred onto a PVDF membrane, and blocked with 5% nonfat milk in Tris-buffered saline for 1 h at room temperature. The membranes were incubated at 4 °C overnight with proper dilution according to the instruments of the following antibodies: GAPDH (#5174, Cell Signaling Technology, Danvers, MA, USA), SIRT1 (#ab32441, Abcam, Cambridgeshire, UK), STAT3 (#9139, Cell Signaling Technology, Danvers, MA, USA), STAT3 phosphorylation (#9145, Cell Signaling Technology, Danvers, MA, USA), OPN (#ab8448, Abcam), and BSP (#ab84787, Abcam, Cambridgeshire, UK). The proteins were visualized using an enhanced chemiluminescence system (Alpha Innotech, San Leandro, CA, USA).
2.8. Bioinformatics Analysis
The microarray dataset was retrieved from EMBL EBI ArrayExpress Data Bank (https://www.ebi.ac.uk/arrayexpress, accessed on 1 July 2020) under Array Express accession E-MTAB-3010 [20]. The histone acetylation modulator proteins (HAMPs) gene family members (n = 73) were defined based on a recent study [11]. The Bioconductor package complexHeatmap was used to visualize the expression pattern of HAMPs during osteoblast differentiation of hDPSCs [21]. GSEA was performed between the Day 21 group and the Day 0 group [22]. The rank is based on the fold change as computed with limma [23]. The database used by GSEA is the Molecular Signatures Database v7.1.
2.9. Statistical Analysis
The quantitative data between two groups were analyzed with Student’s t-test. The two-sided p-value less than 0.05 was defined as statistically significant for the statistical analyses. The data were plotted as mean ± standard deviation (SD). All testing was carried out using Prism 6.0 (GraphPad, San Diego, CA, USA).
3. Results
3.1. A Systematic Landscape of the Expression Pattern of HAMPs during Osteoblast Differentiation of hDPSCs
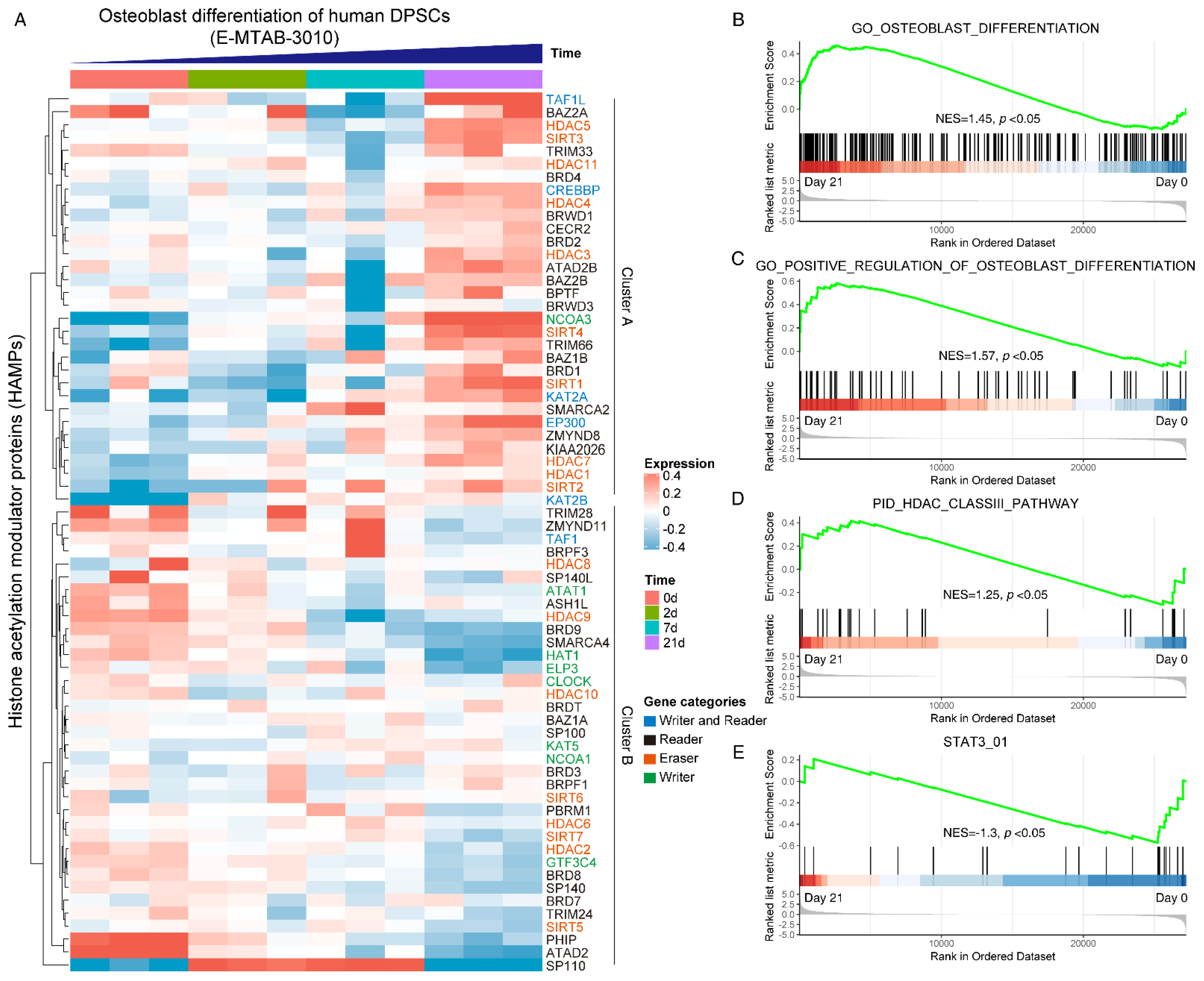
To gain a systematical expression profile of HAMPs during the osteogenic differentiation process of hDPSCs, we analyzed a publicly available transcriptomics dataset using the curated HAMPs gene list. The HAMPs family members were categorized as four clusters, as previously reported [11]. Most HAMP genes were differentially expressed during osteogenic induction, in which two expression clusters can be clearly recognized: Cluster A (TAF1L, HDAC5, NCOA3, SIRT4, SIRT1, etc.) was highly expressed in the Day 21 group, while Cluster B (PHIP, ATAD2, HDAC9, etc.) showed high expression in the Day 0 group (Figure 1A). GSEA pathway analysis displayed an upregulation of the osteoblast differentiation pathway (NES = 1.45) (Figure 1B) and its positive regulation signaling (NES = 1.57) (Figure 1C), validating the successful induction. We noticed that the HDAC class III pathway was upregulated (NES = 1.25) (Figure 1D), whereas the STAT3 signaling was downregulated during osteoblast differentiation (NES = −1.3) (Figure 1E). Taken together, these data indicated that the HAMPs might be involved in osteoblast differentiation and STAT3 might be involved in the pathway.
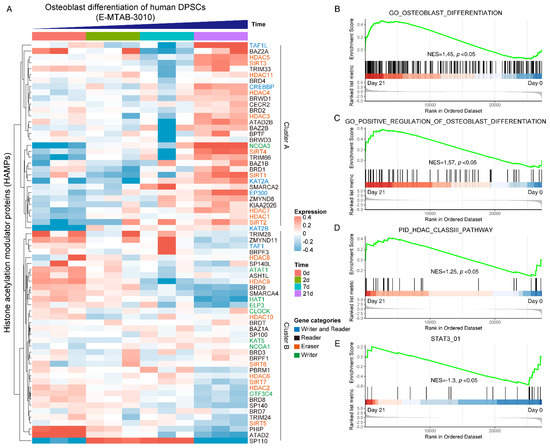
Figure 1.
Systematically analyzing the expression pattern of histone acetylation modulator proteins (HAMPs) during osteoblast differentiation of hDPSCs. (A) Heatmap showing the expression pattern of HAMPs during osteoblast differentiation of hDPSCs. The gene categories of HAMPs are indicated by different colors. The publicly available dataset was under ArrayExpress accession E-MTAB-3010. Red indicates high expression, and blue indicates low expression. (B–E) GSEA analysis of osteoblast differentiation pathway, HDAC class III pathway, and STAT3 pathway on day 0 and day 21. The null hypothesis was “H0: the genes are randomly distributed”.
3.2. SIRT1 Was Upregulated and STAT3 Activation Was Suppressed during Osteogenic Differentiation of hDPSCs
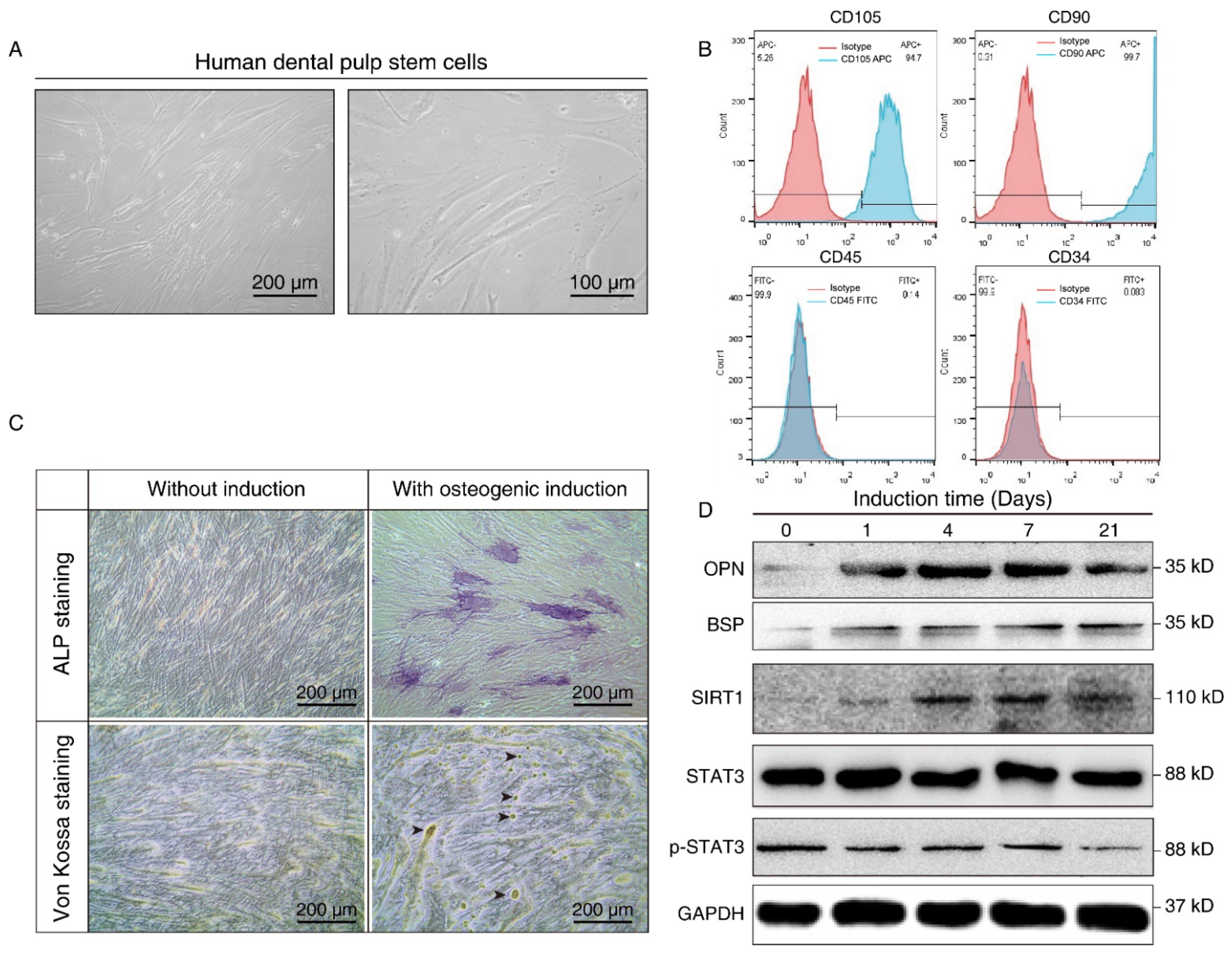
We further isolated and expanded human hDPSCs in vitro. hDPSCs displayed a typical fibroblast-like morphology (Figure 2A). The flow cytometry assay showed that the hDPSCs we isolated were positive for mesenchymal cell surface markers (CD90 and CD105), but were negative for the endothelial-specific marker (CD34) and hematopoietic cell-specific marker (CD45), which validated the immunophenotype of hDPSCs (Figure 2B). Moreover, the osteoblastic phenotype of hDPSCs was analyzed by ALP staining and von Kossa staining. Results showed that osteogenic induction promoted osteogenic differentiation at both the early stage and late stage (Figure 2C). Overall, the above results confirmed the isolation, characterization, and osteogenic differentiation of hDPSCs in vitro.
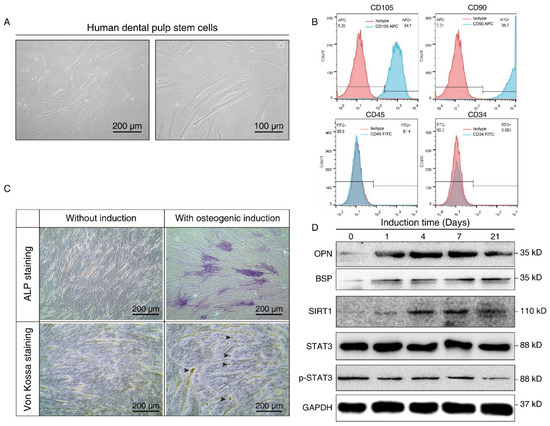
Figure 2.
Isolation, characterization, and osteogenic differentiation of hDPSCs. (A) Morphology of hDPSCs. hDPSCs showed a fibroblast-like morphology after 14 days of osteogenesis. (B) Immunophenotype analysis of the first passage of hDPSCs by flow cytometry. hDPSCs were stained with cell surface marker antigens CD105, CD90, CD45, and CD34. (C) ALP staining (upper panel) and von Kossa staining (lower panel) were performed on day 14 with or without osteogenic induction. Long-term culture of hDPSCs in the differentiation medium showed the capacity to form bone nodules. Arrows indicate osteogenesis. (D) Western blot showing the expression pattern of osteogenesis markers (BSP, OPN), SIRT1, and p-STAT3 at different time points during osteoblast differentiation. OM: Osteogenic induction medium, RSV: Resveratrol.
The differentiated cells at different time points were then subjected to Western blot analysis. Osteopontin (OPN), an early-stage marker in the initial time of osteogenesis, showed the highest expression on days 4 and 7 after induction, whereas bone sialoprotein (BSP), a late-stage osteogenesis marker, increased gradually, and peaked on the 21st day after in vitro differentiation. Consistent with the aforementioned bioinformatics results and the staining results, we demonstrated osteogenic differentiation during osteogenic induction. We also noticed that SIRT1 expression was upregulated after osteogenic induction, while the total-STAT3 expression level did not show a significant change. Nevertheless, p-STAT3 decreased in a time-dependent manner during the process (Figure 2D). It hinted that SIRT1 might also play a role in the phosphorylation of STAT3.
3.3. SIRT1 Counteracted the Activation of STAT3 in hDPSCs
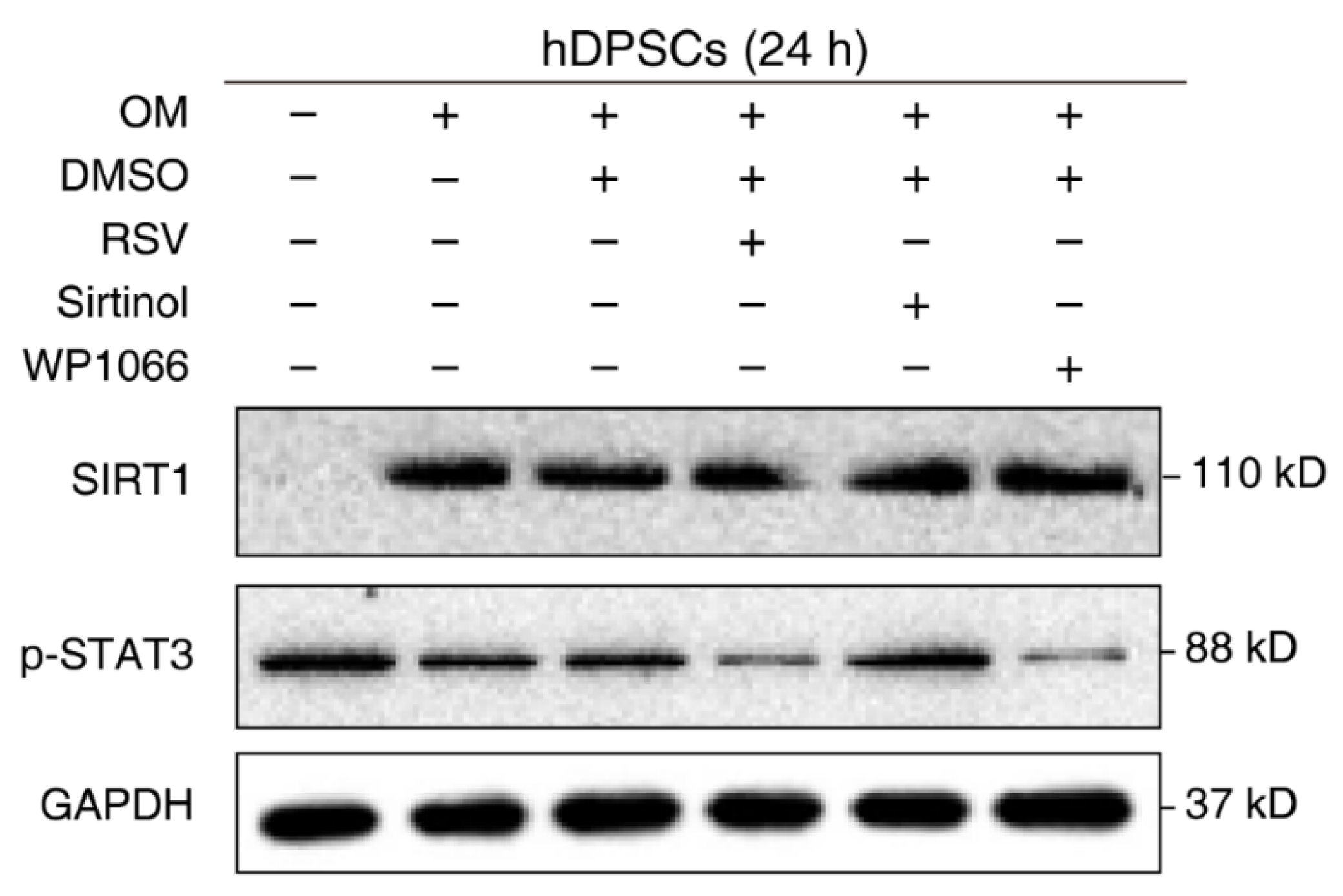
The negative regulation of STAT3 by SIRT1 is well-established in other cell types or disease models. However, their relationship in hDPSCs remains unknown. To verify our previous conjecture and confirm the effect of SIRT1 on STAT3 activation in hDPSCs, cells were treated with the regulatory compounds of SIRT1 and STAT3 and then subjected to Western blot examination. Administration with the SIRT1 inhibitor Sirtinol enhanced STAT3 phosphorylation, while the SIRT1 activator resveratrol (RSV) significantly reduced the phosphorylation of STAT3 at Tyr705. The STAT3 phosphorylation inhibitor WP1066 was used as a positive control for STAT3 repression. Additionally, we used cells with medium and osteogenic medium as a negative control (Figure 3). Together, these results supported the idea that SIRT1 counteracted the activation of STAT3 in hDPSCs.
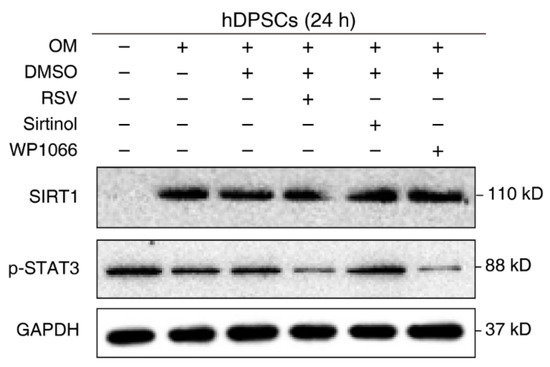
Figure 3.
SIRT1 counteracted the activation of STAT3 in hDPSCs. hDPSCs were treated with the SIRT1 activator (resveratrol, RSV, 200 μM) or inhibitor (sirtinol, 120 μM) for 24 h, then subjected to Western blot examination. A STAT3 phosphorylation inhibitor (WP1066, 5 μM) was used as a positive control for STAT3 inhibition. OM: Osteogenic induction medium, RSV: Resveratrol.
3.4. The SIRT1-STAT3 Axis Regulated the Osteogenic Differentiation of hDPSCs at Both Early and Late Stages
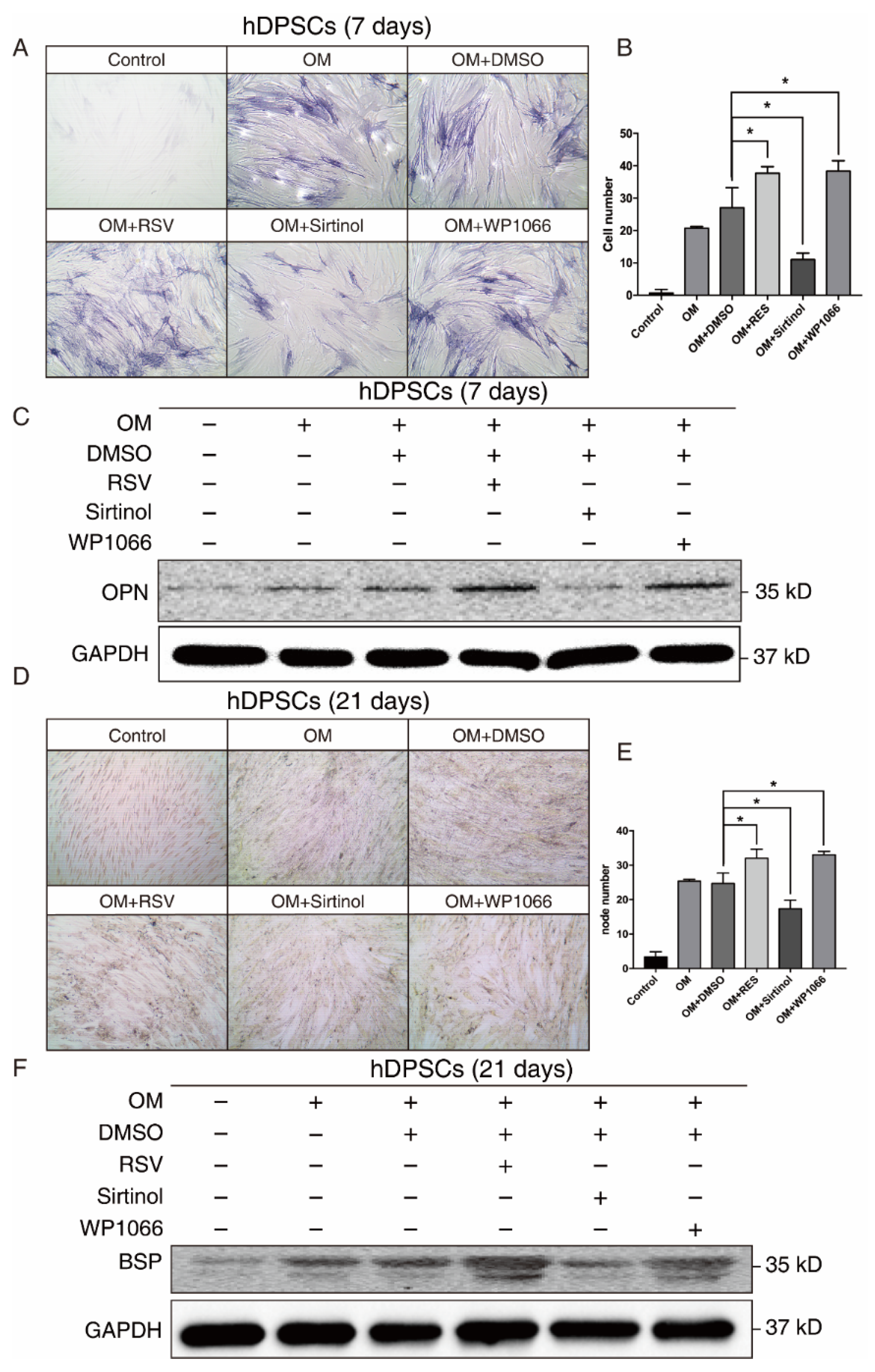
Since both our bioinformatics analysis and in vitro results suggested that SIRT1 and STAT3 may play a crucial role in osteogenic differentiation of hDPSCs, we manipulated the SIRT1-STAT3 signaling pathway, then examined the osteogenic potential of hDPSCs. The SIRT1 activator RSV treatment promoted the osteogenic differentiation of hDPSCs, while sirtinol played an opposite role in the early stage, as measured by ALP staining. The STAT3 inhibitor WP1066 treatment also enhanced osteogenic differentiation of hDPSCs (Figure 4A,B). Besides, the expression level of OPN, an early osteogenesis-associated marker, showed a similar trend to ALP staining (Figure 4C).
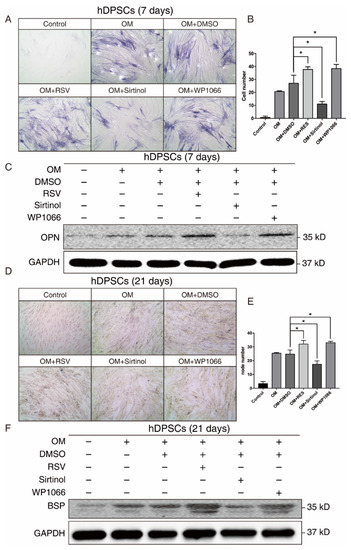
Figure 4.
The SIRT1-STAT3 pathway regulated the osteogenic differentiation of hDPSCs at both early and late stages. (A) ALP staining showing the osteogenic differentiation potential of hDPSCs at day 7 under indicated conditions. (B) Quantitative analysis of the ALP staining activity from (A). The null hypothesis of the quantitative analysis was “H0: there is no difference between two groups”. (C) Western blot showing the expression levels of early osteogenic marker OPN under indicated conditions. (D) Von Kossa staining and the quantitative analysis (E) showing the osteogenic differentiation potential of hDPSCs at day 21 under indicated conditions. The null hypothesis of the quantitative analysis was also “H0: there is no difference between two groups”. (F) Western blot showing the expression levels of late osteogenic marker BSP under indicated conditions. OM: Osteogenic induction medium, RSV: Resveratrol. * p < 0.05.
We further investigated the effect of the SIRT1-STAT3 axis on osteogenic differentiation at the late stage. Both von Kossa staining and Western blot revealed that activating SIRT1 or suppressing the STAT3 signaling pathway significantly improved the osteogenic differentiation of hDPSCs at the late stage (Figure 4D–F). Collectively, these findings demonstrated that the SIRT1-STAT3 axis was critically involved in the osteogenic differentiation of hDPSCs, in which activation of SIRT1 enhanced the osteogenic differentiation potential via the repression of STAT3 at both early and late stages.
4. Discussion
hDPSCs are one of the up-and-coming candidate cells in the tissue engineering field to repair dental pulp injury or bone tissue defect due to their ability of self-renewal and multiple differentiation [4]. It is the core step to make clear the mechanisms of hDPSCs’ osteogenic differentiation to acquire predictable bone regeneration. In this study, we firstly performed a systematical analysis of the expression pattern of HAMPs during osteoblast differentiation of hDPSCs, and found that the HDAC class III pathway was upregulated, whereas the STAT3 signaling was downregulated during osteoblast differentiation using a publicly available time-course dataset. We delineated that SIRT1, a class III HDAC, contributes to osteogenic differentiation of hDPSCs through counteracting STAT3 activation.
More and more attention has been paid to epigenetic regulation on hDPSCs, as epigenetics could modulate cellular functions without entailing a change in the DNA sequence [24,25]. Several previous studies have suggested a linkage between epigenetic reprogramming and hDPSCs’ differentiation, indicating that epigenetic regulation plays an important role in mineralized regeneration [26,27,28,29]. It has been observed that MI192, which is the selective inhibitor of HDAC2 and 3, promotes the osteogenic differentiation of hDPSCs [30]. The HDAC inhibitors trichostatin A (TSA) and valproic acid (VPA) promoted hDPSCs’ proliferation, cell migration, and adhesion [31]. HDAC2 silencing resulted in an increasing of mineralization markers OPN and BSP, and enhanced osteogenic differentiation, resembling the effect of VPA [32]. Here, we found that the SIRT1 activator promoted the osteogenic differentiation of hDPSCs. Resveratrol, a natural activator of SIRT1, was reported to activate SIRT1 by generating a structural change that could better attach to the coumarin group, which has already entered into clinical trials [33,34,35]. Our study implicated the potential utility of resveratrol in inducing osteogenic differentiation.
SIRT1 plays a role in regulating the activation of STAT3 in a variety of processes. It has been described that SIRT1 downregulates STAT3 phosphorylation directly [36,37,38]. Since STAT3 plays a leading role in inflammation, it seems reasonable to hypothesize the interactions between the inflammatory process and osteogenic differentiation. Further work is required to investigate the crosstalk between pulpal inflammation and hDPSCs’ osteogenic differentiation.
There are several limitations of this study. First, we manipulated the activity of the SIRT1-STAT3 signaling pathway through small-molecule compounds. The conclusion can be further strengthened by the knock-out and overexpression of the target genes. Second, this study was mainly conducted in silico and in vitro. In vivo research and animal model research need to be carried out to explore the clinical potential of hDPSCs in regenerative medicine. Third, we identified that SIRT1 promotes osteogenic differentiation through repressing STAT3. However, the detailed molecular mechanisms underlying STAT3 and hDPSCs warrant further investigation.
5. Conclusions
In summary, we performed the first comprehensive analysis of the expression pattern of HAMPs during osteoblast differentiation of hDPSCs. We found that SIRT1 was upregulated and STAT3 activation was suppressed during osteogenic differentiation. SIRT1 counteracted the activation of STAT3 to promote osteogenic differentiation of hDPSCs at both early and late stages, which highlights the promising utility of SIRT1 activators in hDPSCs-based therapies for bone augmentation strategies. Further studies are needed to fully clarify the roles of other HAMPs in the osteogenic differentiation of hDPSCs and underlying mechanisms.
Author Contributions
Conceptualization: D.Z. and S.X.; methodology: D.Z., W.K. and Y.W.; data curation: D.Z. and J.H.; formal analysis: D.Z. and J.Y.; investigation: J.G., W.Y. and X.T.; writing—original draft preparation: D.Z. and S.X.; writing—review and editing: D.Z., X.T., J.G. and S.X.; funding acquisition: J.G. and S.X. All authors gave final approval and agree to be accountable for all aspects of the work. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81801401, 82001111), and Jiangsu Provincial Medical Youth Talent (QNRC2016119).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the Institutional Review Board of Nanjing Stomatological Hospital (KY-2020NL-029).
Informed Consent Statement
Informed consent was obtained from every study participant prior to study initiation.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
hDPSCs: human dental pulp stem cells; HAMPs: histone acetylation modulator proteins; SIRT1: sirtuin-1; STAT3: signal transducer and activator of transcription 3; BSP: bone sialoprotein; HDACs: histone deacetylases; HATs: histone acetyltransferases; OPN: osteopontin; ALP: alkaline phosphatase; OM: osteogenic induction medium; RSV: resveratrol.
References
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [Green Version]
- La Noce, M.; Pain, F.; Spina, A.; Naddeo, P.; Montella, R.; Desiderio, V.; De Rosa, A.; Papaccio, G.; Tirino, V.; Laino, L. Dental pulp stem cells: State of the art and suggestions for a true translation of research into therapy. J. Dent. 2014, 42, 761–768. [Google Scholar] [CrossRef] [Green Version]
- Aydin, S.; Sahin, F. Stem cells derived from dental tissues. Adv. Exp. Med. Biol. 2019, 1144, 123–132. [Google Scholar]
- Yamada, Y.; Nakamura-Yamada, S.; Kusano, K.; Baba, S. Clinical potential and current progress of dental pulp stem cells for various systemic diseases in regenerative medicine: A concise review. Int. J. Mol. Sci. 2019, 20, 1132. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Lee, J.H.; Bae, J.; Bu, Y.; Kim, E.C. Human dental pulp stem cells are more effective than human bone marrow-derived mesenchymal stem cells in cerebral ischemic injury. Cell Transpl. 2017, 26, 1001–1016. [Google Scholar] [CrossRef]
- Awais, S.; Balouch, S.S.; Riaz, N.; Choudhery, M.S. Human dental pulp stem cells exhibit osteogenic differentiation potential. Open Life Sci. 2020, 15, 229–236. [Google Scholar] [CrossRef]
- Stanko, P.; Kaiserova, K.; Altanerova, V.; Altaner, C. Comparison of human mesenchymal stem cells derived from dental pulp, bone marrow, adipose tissue, and umbilical cord tissue by gene expression. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2014, 158, 373–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, H.F.; Smith, A.J.; Fleming, G.J.P.; Cooper, P.R. Epigenetic modulation of dental pulp stem cells: Implications for regenerative endodontics. Int. Endod. J. 2016, 49, 431–446. [Google Scholar] [CrossRef]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Zhou, J.; Jiang, J.; Yuan, J.; Zhang, Y.; Wei, X.; Loo, N.; Wang, Y.; Pan, Y.; Zhang, T.; et al. Genomic characterization of genes encoding histone acetylation modulator proteins identifies therapeutic targets for cancer treatment. Nat. Commun. 2019, 10, 733. [Google Scholar] [CrossRef]
- Javaid, N.; Choi, S. Acetylation- and methylation-related epigenetic proteins in the context of their targets. Genes 2017, 8, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, Y.; Cooper, P.R.; Shimizu, E.; Kobayashi, Y.; Smith, A.J.; Duncan, H.F. Histone acetylation as a regenerative target in the dentine-pulp complex. Front. Genet. 2020, 11, 1. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, M.; Ge, Y.; Wang, X. SIRT1 and aging related signaling pathways. Mech. Ageing Dev. 2020, 187, 111215. [Google Scholar] [CrossRef]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watroba, M.; Dudek, I.; Skoda, M.; Stangret, A.; Rzodkiewicz, P.; Szukiewicz, D. Sirtuins, epigenetics and longevity. Ageing Res. Rev. 2017, 40, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, F.; Xu, Y.; Wei, J.; Zhang, Y.; Yang, H.; Gao, B.; Yu, G.; Fang, D. JAK1-mediated Sirt1 phosphorylation functions as a negative feedback of the JAK1-STAT3 pathway. J. Biol. Chem. 2018, 293, 11067–11075. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Xu, J.; Xiong, X.; Deng, Y. Salidroside inhibits MAPK, NF-kappaB, and STAT3 pathways in psoriasis-associated oxidative stress via SIRT1 activation. Redox Rep. 2019, 24, 70–74. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; You, S.; Tian, Y.; Lu, S.; Cao, L.; Sun, Y.; Zhang, N. WWP2 regulates SIRT1-STAT3 acetylation and phosphorylation involved in hypertensive angiopathy. J. Cell Mol. Med. 2020, 24, 9041–9054. [Google Scholar] [CrossRef]
- Dernowsek, J.A.; Pereira, M.C.; Fornari, T.A.; Macedo, C.; Assis, A.F.; Donate, P.B.; Bombonato-Prado, K.F.; Passos-Bueno, M.R.; Passos, G.A. Posttranscriptional interaction between miR-450a-5p and miR-28-5p and STAT1 mRNA triggers osteoblastic differentiation of human mesenchymal stem cells. J. Cell. Biochem. 2017, 118, 4045–4062. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: Berlin/Heidelberg, Germany, 2005; pp. 397–420. [Google Scholar]
- Ozkul, Y.; Galderisi, U. The impact of epigenetics on mesenchymal stem cell biology. J. Cell Physiol. 2016, 231, 2393–2401. [Google Scholar] [CrossRef]
- Huynh, N.C.-N.; Everts, V.; Ampornaramveth, R.S. Histone deacetylases and their roles in mineralized tissue regeneration. Bone Rep. 2017, 7, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.S.; Liang, J.P.; Wang, J.; Liu, B. Histone acetylation regulates osteodifferentiation of hDPSCs via DSPP. Front. Biosci.-Landmark 2013, 18, 1072–1079. [Google Scholar]
- Zhou, D.; Gan, L.; Peng, Y.; Zhou, Y.; Zhou, X.; Wan, M.; Fan, Y.; Xu, X.; Zhou, X.; Zheng, L.; et al. Epigenetic regulation of dental pulp stem cell fate. Stem. Cells Int. 2020, 2020, 8876265. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Lian, W.S.; Kuo, C.W.; Ke, H.J.; Wang, S.Y.; Kuo, P.C.; Jahr, H.; Wang, F.S. Epigenetic regulation of skeletal tissue integrity and osteoporosis development. Int. J. Mol. Sci. 2020, 21, 4923. [Google Scholar] [CrossRef]
- Zhang, H.; Ji, L.; Yang, Y.; Zhang, X.; Gang, Y.; Bai, L. The role of HDACs and HDACi in cartilage and osteoarthritis. Front. Cell Dev. Biol. 2020, 8, 560117. [Google Scholar] [CrossRef] [PubMed]
- Man, K.; Lawlor, L.; Jiang, L.H.; Yang, X.B. The selective histone deacetylase inhibitor mi192 enhances the osteogenic differentiation efficacy of human dental pulp stromal cells. Int. J. Mol. Sci. 2021, 22, 5224. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, Z.; He, X.; Liu, N.; Liu, B.; Sun, L.; Wang, J.; Ma, F.; Duncan, H.; He, W.; et al. Effects of histone deacetylase inhibitors on regenerative cell responses in human dental pulp cells. Int. Endod. J. 2018, 51, 767–778. [Google Scholar] [CrossRef]
- Paino, F.; La Noce, M.; Tirino, V.; Naddeo, P.; Desiderio, V.; Pirozzi, G.; De Rosa, A.; Laino, L.; Altucci, L.; Papaccio, G. Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: Evidence for HDAC2 involvement. Stem. Cells. 2014, 32, 279–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, A.Y.; Motechin, R.A.; Wiesenfeld, M.Y.; Holz, M.K. The therapeutic potential of resveratrol: A review of clinical trials. NPJ Precis. Oncol. 2017, 1, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquariello, R.; Verdile, N.; Brevini, T.A.L.; Gandolfi, F.; Boiti, C.; Zerani, M.; Maranesi, M. The role of resveratrol in mammalian reproduction. Molecules 2020, 25, 4554. [Google Scholar] [CrossRef]
- Sarubbo, F.; Esteban, S.; Miralles, A.; Moranta, D. Effects of resveratrol and other polyphenols on Sirt1: Relevance to brain function during aging. Curr. Neuropharmacol. 2018, 16, 126–136. [Google Scholar] [CrossRef]
- Nie, Y.Z.; Erion, D.M.; Yuan, Z.L.; Dietrich, M.; Shulman, G.I.; Horvath, T.L.; Gao, Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol. 2009, 11, 492–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Li, Y.; Lou, M.; Xia, W.; Liu, Q.; Xie, G.; Liu, L.; Liu, B.; Yang, J.; Qin, M. Baicalin regulates SirT1/STAT3 pathway and restrains excessive hepatic glucose production. Pharmacol. Res. 2018, 136, 62–73. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Y.; Huang, S.; Deng, C.; Zhou, S.; Yang, J.; Cao, Y.; Xu, L.; Yuan, Y.; Yang, J.; et al. SIRT1 inhibits gastric cancer proliferation and metastasis via STAT3/MMP-13 signaling. J. Cell Physiol. 2019, 234, 15395–15406. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).