Mitophagy in Yeast: Decades of Research
Abstract
:1. Introduction
2. Yeast as a Study Model
3. Mitophagy in Yeast
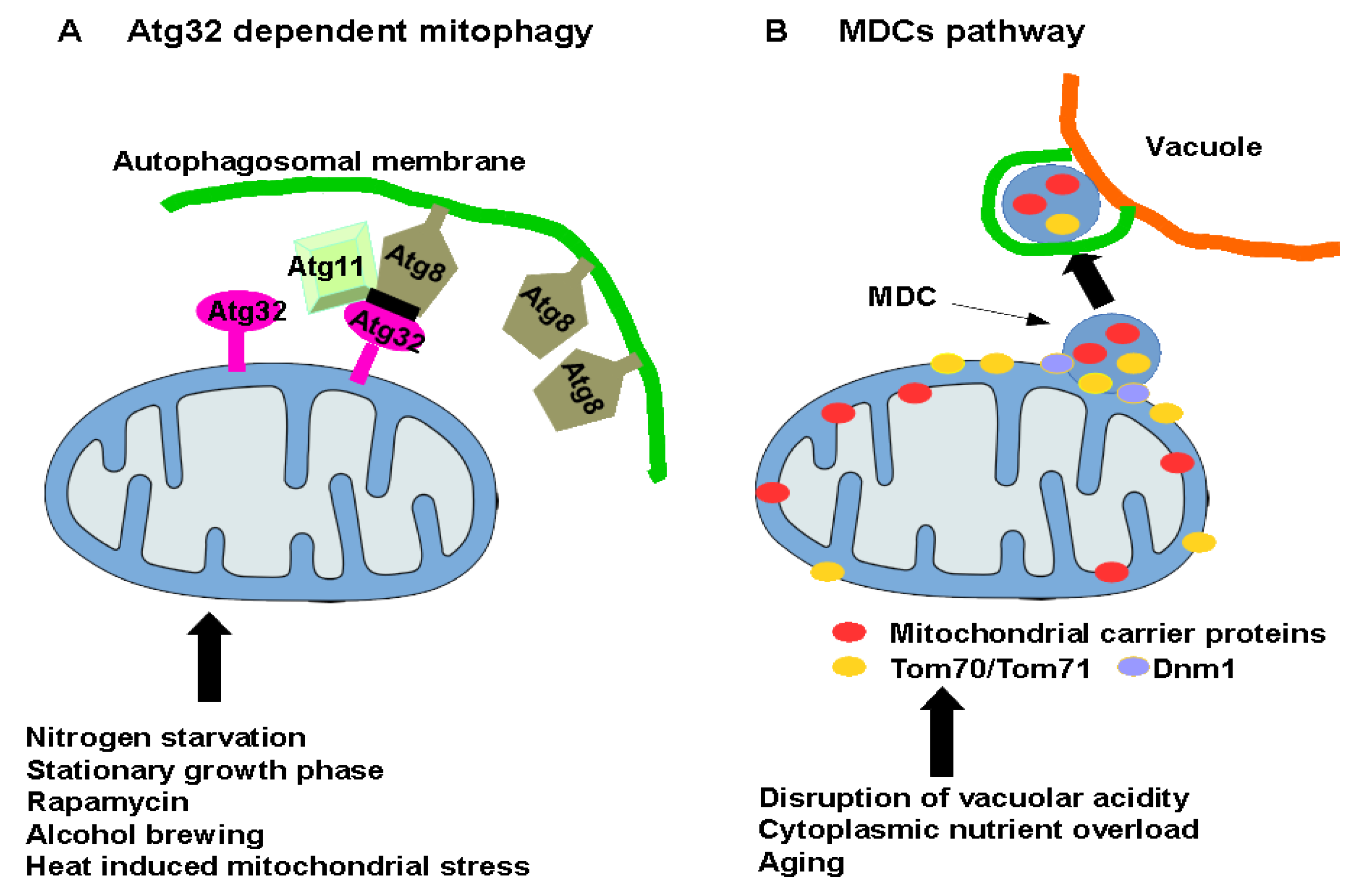
4. Key Role of the Atg32 Protein
5. An Unconventional Pathway for Degradation of Mitochondria in Yeast
6. Induction of Mitophagy in S. cerevisiae
7. Are ROS the Main Culprits?
8. Factors Modulating Mitophagy
9. Physiological Role of Mitophagy in S. cerevisiae
10. Mitophagy in Other Yeast Species
11. Yeast Saccharomyces cerevisiae Could Be Used to Express Human Proteins
12. Future Directions
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMBRA1 | autophagy and beclin 1 regulator 1 |
| BNIP3 | BCL2 interacting protein 3 |
| BNIP3L/NIX | BCL2 interacting protein 3 like |
| CCCP | Carbonyl cyanide m-chlorophénylhydrazone |
| CK2 | casein kinase 2 |
| DHFR | Dihydrofolate reductase |
| FCCP | Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone |
| FKBP8 | FKBP prolyl isomerase 8 |
| FUNDC1 | FUN14 domain containing 1 |
| GFP | green fluorescent protein |
| GSH | reduced glutathione |
| LIR | LC3-interacting region |
| MAM | mitochondria associated membrane |
| NAC | N-acetyl cysteine |
| PHB2 | prohibitin 2 |
| PINK1 | PTEN induced kinase 1 |
| RFP | red fluorescent protein |
References
- Chance, B. The respiratory chain and oxidative phosphorylation. Adv. Enzymol. Relat. Subj. Biochem. 1956, 17, 65. [Google Scholar]
- Lehninger, A.L.; Wadkins, C.L. Oxidative phosphorylation. Annu. Rev. Biochem. 1962, 31, 47–78. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Moyle, J. Chemiosmotic hypothesis of oxidative phosphorylation. Nature 1967, 213, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Hockenbery, D.M.; Oltvai, Z.N.; Yin, X.M.; Milliman, C.; Korsmeyer, S.J. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell 1993, 5, 241–251. [Google Scholar] [CrossRef]
- Loschen, G.; Flohe, L.; Chance, B. Respiratory chain linked H2O2 production in pigeon heart mitochondria. FEBS Lett. 1972, 18, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Boveris, A.; Chance, B. The mitochondria1 generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J. 1973, 134, 707–716. [Google Scholar] [CrossRef]
- Boveris, A. Mitochondrial production of superoxide radicals and hydrogen peroxide. Adv. Exp. Med. Biol. 1977, 53, 382–393. [Google Scholar]
- Forman, H.J.; Boveris, A. Superoxide Radical and Hydrogen Peroxide in Mitochondria. In Free Radicals in Biology; Pryor, W.A., Ed.; Academic Press: New York, NY, USA, 1982; Volume V, pp. 65–90. [Google Scholar]
- Lennicke, C.; Cochemé, H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef]
- Shields, H.J.; Traa, A.; Van Raamsdonk, J.M. Beneficial and Detrimental Effects of Reactive Oxygen Species on Lifespan: A Comprehensive Review of Comparative and Experimental Studies. Front. Cell Dev. Biol. 2021, 9, 628157. [Google Scholar] [CrossRef] [PubMed]
- Barja, G. Updating the mitochondrial free radical theory of aging: An integrated view, key aspects, and confounding concepts. Antioxid. Redox Signal. 2013, 19, 1420–1445. [Google Scholar] [CrossRef] [Green Version]
- Salmon, A.B.; Richardson, A.; Perez, V.I. Update on the oxidative stress theory of aging: Does oxidative stress play a role in aging or healthy aging? Free Radic. Biol. Med. 2010, 48, 642–655. [Google Scholar] [CrossRef] [Green Version]
- Voos, W.; Rottgers, K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta 2002, 1592, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Deshwal, S.; Fiedler, K.U.; Langer, T. Mitochondrial proteases: Multifaceted regulators of mitochondrial plasticity. Annu. Rev. Biochem. 2020, 89, 501–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quiros, P.M.; Langer, T.; Lopez-Otin, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015, 16, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Livnat-Levanon, N.; Glickman, M.H. Ubiquitin-proteasome system and mitochondria—Reciprocity. Biochim. Biophys. Acta 2011, 1809, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Soubannier, V.; McLelland, G.-L.; Zunino, R.; Braschi, E.; Rippstein, P.; Fon, E.A.; McBride, H.M. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 2012, 22, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Soubannier, V.; Rippstein, P.; Kaufman, B.A.; Shoubridge, E.A.; McBride, H.M. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS ONE 2012, 7, e52830. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, A.; McLelland, G.L.; Fon, E.A.; McBride, H.M. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014, 33, 2142–2156. [Google Scholar] [CrossRef] [Green Version]
- Vasam, G.; Nadeau, R.; Cadete, V.J.J.; Lavallée-Adam, M.; Menzies, K.J.; Burelle, Y. Proteomics characterization of mitochondrial-derived vesicles under oxidative stress. FASEB J 2021, 35, e21278. [Google Scholar] [CrossRef]
- Ashford, T.P.; Porter, K.R. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 1962, 12, 198–202. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Pedro, J.M.B.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef] [PubMed]
- Lemasters, J.J. Perspective-Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuv. Res. 2005, 8, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Mortimer, R.K.; Johnston, J.R. Genealogy of principal strains of the yeast genetic stock center. Genetics 1986, 113, 35–43. [Google Scholar] [CrossRef]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Ralser, M.; Kuhl, H.; Ralser, M.; Werber, M.; Lehrach, H.; Breitenbach, M.; Timmermann, B. The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: Insights into ancestry and physiology of a laboratory mutt. Open Biol. 2012, 2, 120093. [Google Scholar] [CrossRef] [Green Version]
- Gaisne, M.; Becam, A.M.; Verdiere, J.; Herbert, C.J. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 1999, 36, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Kolitsida, P.; Zhou, J.; Rackiewicz, M.; Nolic, V.; Dengjel, J.; Abeliovich, H. Phosphorylation of mitochondrial matrix proteins regulates their selective mitophagic degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 20517–20527. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.L.; Thorsness, P.E. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 1998, 111, 2455–2464. [Google Scholar] [CrossRef] [PubMed]
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992, 119, 301–311. [Google Scholar] [CrossRef] [Green Version]
- Noda, T.; Matsuura, A.; Wada, Y.; Ohsumi, Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995, 210, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.W.; Zubenko, G.S.; Parker, R.R. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics 1982, 102, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Kissova, I.; Deffieu, M.; Manon, S.; Camougrand, N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004, 279, 39068–39074. [Google Scholar] [CrossRef] [Green Version]
- Velours, G.; Boucheron, C.; Manon, S.; Camougrand, N. Dual cell wall/mitochondria localization of the ‘SUN’ family proteins. FEMS Microbiol. Lett. 2002, 207, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Welter, E.; Montino, M.; Reinhold, R.; Schlotterhose, P.; Krick, R.; Dudek, J.; Rehling, P.; Thumm, M. Uth1 is a mitochondrial inner membrane protein dispensable for post- log-phase and rapamycin-induced mitophagy. FEBS J. 2013, 280, 4970–4982. [Google Scholar] [CrossRef] [Green Version]
- Kissova, I.; Salin, B.; Schaeffer, J.; Bhatia, S.; Manon, S.; Camougrand, N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 2007, 3, 329–336. [Google Scholar] [CrossRef] [Green Version]
- Priault, M.; Salin, B.; Schaeffer, J.; Vallette, F.M.; di Rago, J.P.; Martinou, J.C. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005, 12, 1613–1621. [Google Scholar] [CrossRef] [Green Version]
- Ogier-Denis, E.; Codogno, P. Autophagy: A barrier or an adaptive response to cancer. Biochim. Biophys. Acta 2003, 1603, 113–128. [Google Scholar] [CrossRef]
- Nowikovsky, K.; Reipert, S.; Devenish, R.J.; Schweyen, R.J. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007, 14, 1647–1656. [Google Scholar] [CrossRef]
- Tal, R.; Winter, G.; Ecker, N.; Klionsky, D.J.; Abeliovich, H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 2007, 282, 5617–5624. [Google Scholar] [CrossRef] [Green Version]
- Journo, D.; Mor, A.; Abeliovich, H. Aup1-mediated regulation of Rtg3 during mitophagy. J. Biol. Chem. 2009, 284, 35885–35895. [Google Scholar] [CrossRef] [Green Version]
- Kanki, T.; Klionsky, D.J. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008, 283, 32386–32393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanki, T.; Wang, K.; Baba, M.; Bartholomew, C.R.; Lynch-Day, M.A.; Du, Z.; Geng, J.; Mao, K.; Yang, Z.; Yen, W.-L.; et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 2009, 20, 4730–4738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, K.; Kondo-Okamoto, N.; Ohsumi, Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 2009, 17, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Kanki, T.; Wang, K.; Cao, Y.; Baba, M.; Klionsky, D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009, 17, 98–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aihara, M.; Jin, X.; Kurihara, Y.; Yoshida, Y.; Matsushima, Y.; Oku, M.; Hirota, Y.; Saigusa, T.; Aoki, Y.; Uchiumi, T.; et al. The Tor and the Sin3-Rpd3 complex regulate expression of the mitophagy receptor protein Atg32 in yeast. J. Cell Sci. 2014, 127, 3184–3196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Shu, W.; Li, Y.-M.; Mari, M.; Yan, C.; Wang, D.; Yin, Z.-H.; Jiang, W.; Zhou, Y.; Okamoto, K.; et al. The Paf1 complex transcriptionally regulates the mitochondrial-anchored protein Atg32 leading to activation of mitophagy. Autophagy 2020, 16, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Eiyama, A.; Okamoto, K. Protein N-terminal Acetylation by the NatA Complex Is Critical for Selective Mitochondrial Degradation. J. Biol. Chem. 2015, 290, 25034–25044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakakibara, K.; Eiyama, A.; Suzuki, S.W.; Sakoh-Nakatogawa, M.; Okumura, N.; Tani, M.; Hashimoto, A.; Nagumo, S.; Kondo-Okamoto, N.; Kondo-Kakuta, C.; et al. Phospholipid methylation controls Atg32-mediated mitophagy and Atg8 recycling. EMBO J. 2015, 34, 2703–2719. [Google Scholar] [CrossRef] [Green Version]
- Kubota, M.; Okamoto, K. The protein N-terminal acetyltransferase A complex contributes to yeast mitophagy via promoting expression and phosphorylation of Atg32. J. Biochem. 2021, 170, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Kanki, T.; Hirota, Y.; Kurihara, Y.; Saigusa, T.; Uchiumi, T.; Kang, D. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 2011, 22, 3206–3217. [Google Scholar] [CrossRef]
- Kanki, T.; Kurihara, Y.; Jin, X.; Goda, T.; Ono, Y.; Aihara, M.; Hirota, Y.; Saigusa, T.; Aoki, Y.; Uchiumi, T.; et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013, 14, 788–794. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, K.; Fukuda, T.; Yamashita, S.I.; Saigusa, T.; Kurihara, Y.; Yoshida, Y.; Kirisako, H.; Nakatogawa, H.; Kanki, T. The PP2A-like Protein Phosphatase Ppg1 and the Far Complex Cooperatively Counteract CK2-Mediated Phosphorylation of Atg32 to Inhibit Mitophagy. Cell Rep. 2018, 23, 3579–3590. [Google Scholar] [CrossRef]
- Wang, K.; Jin, M.; Liu, X.; Klionsky, D.J. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy 2013, 9, 1828–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.; Katzenell, S.; Reinhart, E.F.; Bauer, K.M.; Pellegrini, M.; Ragusa, M.J. A pseudo-receiver domain in Atg32 is required for mitophagy. Autophagy 2018, 14, 1620–1628. [Google Scholar] [CrossRef] [Green Version]
- Camougrand, N.; Vigié, P.; Gonzalez, C.; Manon, S.; Bhatia-Kiššová, I. The yeast mitophagy receptor Atg32 is ubiquitinated and degraded by the proteasome. PLoS ONE 2020, 15, e0241576. [Google Scholar] [CrossRef]
- Levchenko, M.; Lorenzi, I.; Dudek, J. The Degradation Pathway of the Mitophagy Receptor Atg32 Is Re-Routed by a Posttranslational Modification. PLoS ONE 2016, 11, e0168518. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Hughes, C.E.; Henderson, K.A.; Yazvenko, N.; Gottschling, D.E. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife 2016, 5, e13943. [Google Scholar] [CrossRef] [PubMed]
- Scheckhuber, C.Q.; Erjavec, N.; Tinazli, A.; Hamann, A.; Nyström, T.; Osiewacz, H.D. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007, 9, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Kanki, T.; Klionsky, D.J.; Okamoto, K. Mitochondria autophagy in yeast. Antioxid Redox Signal. 2011, 14, 1989–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmieri, F.; Pierri, C.L. Structure and function of mitochondrial carriers—Role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 2010, 584, 1931–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.L.; Gottschling, D.E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 2012, 492, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Wellen, K.E.; Thompson, C.B. Cellular metabolic stress: Considering how cells respond to nutrient excess. Mol. Cell. 2010, 40, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Vigié, P.; Cougouilles, E.; Bhatia-Kiššová, I.; Salin, B.; Blancard, C.; Camougrand, N. The mitochondrial phosphatidylserine decarboxylase Psd1 is involved in nitrogen starvation-induced mitophagy in yeast. J. Cell Sci. 2019, 132, jcs221655. [Google Scholar] [CrossRef] [Green Version]
- Ano, Y.; Hattori, T.; Kato, N.; Sakai, Y. Intracellular ATP correlates with mode of pexophagy in Pichia pastoris. Biosci. Biotechnol. Biochem. 2005, 69, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Deffieu, M.; Bhatia-Kissová, I.; Salin, B.; Galinier, A.; Manon, S.; Camougrand, N. Glutathione participates in the regulation of mitophagy in yeast. J. Biol. Chem. 2009, 284, 14828–14837. [Google Scholar] [CrossRef] [Green Version]
- Grant, C.M.; Dawes, I.W. Synthesis and role of glutathione in protection against oxidative stress in yeast. Redox Rep. 1996, 2, 223–229. [Google Scholar] [CrossRef]
- Huang, Y.J.; Klionsky, D.J. Yeast mitophagy: Unanswered questions. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129932. [Google Scholar] [CrossRef] [PubMed]
- Deffieu, M.; Bhatia-Kiššová, I.; Salin, B.; Klionsky, D.J.; Pinson, B.; Manon, S.; Camougrand, N. Increased levels of reduced cytochrome b and mitophagy components are required to trigger nonspecific autophagy following induced mitochondrial dysfunction. J. Cell Sci. 2013, 126, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Mao, K.; Wang, K.; Zhao, M.; Xu, T.; Klionsky, D.J. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 2011, 193, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Mendl, N.; Occhipinti, A.; Müller, M.; Wild, P.; Dikic, I.; Reichert, A.S. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 2011, 124, 1339–1350. [Google Scholar] [CrossRef] [Green Version]
- Mao, K.; Wang, K.; Liu, X.; Klionsky, D.J. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell. 2013, 26, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, M.; Reichert, A.S. Mitophagy, mitochondrial dynamics and the general stress response in yeast. Biochem. Soc. Trans. 2011, 39, 1514–1519. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, V.; Medeiros, T.C.; Vilaça, R.; Pereira, A.T.; Chaves, S.R.; Côrte-Real, M.; Moradas-Ferreira, P.; Costa, V. Ceramide signalling impinges on Sit4p and Hog1p to promote mitochondrial fission and mitophagy in Isc1p-deficient cells. Cell Signal. 2015, 27, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.-I.; Jin, X.; Furukawa, K.; Hamasaki, M.; Nezu, A.; Otera, H.; Saigusa, T.; Yoshimori, T.; Sakai, Y.; Mihara, K.; et al. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 2016, 215, 649–665. [Google Scholar] [CrossRef]
- Kumar, R.; Rahman, M.A.; Nazarko, T.Y. Nitrogen Starvation and Stationary Phase Lipophagy Have Distinct Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 9094. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.L., Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 1957, 3, 349–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karavaeva, I.E.; Golyshev, S.A.; Smirnova, E.A.; Sokolov, S.S.; Severin, F.F.; Knorre, D.A. Mitochondrial depolarization in yeast zygotes inhibits clonal expansion of selfish mtDNA. J. Cell Sci. 2017, 130, 1274–1284. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, Y.; Kanki, T.; Aoki, Y.; Hirota, Y.; Saigusa, T.; Uchiumi, T.; Kang, D. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 2012, 287, 3265–3272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin-Umer, M.A.; McLaughlin, J.E.; Butterly, M.S.; McCormick, S.; Tumer, N.E. Elimination of damaged mitochondria through mitophagy reduces mitochondrial oxidative stress and increases tolerance to trichothecenes. Proc. Natl. Acad. Sci. USA 2014, 111, 11798–11803. [Google Scholar] [CrossRef] [Green Version]
- Richard, V.R.; Leonov, A.; Beach, A.; Burstein, M.T.; Koupaki, O.; Gomez-Perez, A.; Levy, S.; Pluska, L.; Mattie, S.; Rafeh, R. Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis. Aging 2013, 5, 234–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiroma, S.; Jayakody, L.N.; Horie, K.; Okamoto, K.; Kitagaki, H. Enhancement of ethanol fermentation in Saccharomyces cerevisiae sake yeast by disrupting mitophagy function. Appl. Environ. Microbiol. 2014, 80, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Jing, H.; Liu, H.; Lu, Z.; Liuqing, C.; Tan, X. Mitophagy Improves Ethanol Tolerance in Yeast: Regulation by Mitochondrial Reactive Oxygen Species in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 2020, 30, 1876–1884. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Goldsmith, J.; Tankka, A.; Bustamante Eguiguren, S.; Gimenez, A.A.; Vick, L.; Debnath, J.; Vlahakis, A. Atg32-dependent mitophagy sustains spermidine and nitric oxide required for heat-stress tolerance in Saccharomyces cerevisiae. J. Cell Sci. 2021, 134, jcs253781. [Google Scholar] [CrossRef]
- Nagi, M.; Tanabe, K.; Nakayama, H.; Ueno, K.; Yamagoe, S.; Umeyama, T.; Ohno, H.; Miyazaki, Y. Iron-depletion promotes mitophagy to maintain mitochondrial integrity in pathogenic yeast Candida glabrata. Autophagy 2016, 12, 1259–1271. [Google Scholar] [CrossRef] [Green Version]
- Hoshida, H.; Kagawa, S.; Ogami, K.; Akada, R. Anoxia-induced mitophagy in the yeast Kluyveromyces marxianus. FEMS Yeast Res. 2020, 20, foaa057. [Google Scholar] [CrossRef]
- Hoffman, C.S.; Wood, V.; Fantes, P.A. An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System. Genetics 2015, 201, 403–423. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.L.; Li, M.; Suo, F.; Liu, X.M.; Shen, E.Z.; Yang, B.; Dong, M.Q.; He, W.Z.; Du, L.L. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 2013, 9, e1003715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Yoshida, T.; Kikuchi, S.; Nagao, K.; Kokubu, A.; Pluskal, T.; Villar-Briones, A.; Nakamura, T.; Yanagida, M. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 3540–3545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Liu, X.M.; Yu, Z.Q.; Sun, L.L.; Xiong, X.; Dong, M.Q.; Li-Lin Du, L.L. Atg20- and Atg24-family proteins promote organelle autophagy in fission yeast. J. Cell Sci. 2016, 129, 4289–4304. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, T.; Yuki Ebi, Y.; Saigusa, T.; Furukawa, K.; Yamashita, S.I.; Inoue, K.; Kobayashi, D.; Yoshida, Y.; Kanki, T. Atg43 tethers isolation membranes to mitochondria to promote starvation-induced mitophagy in fission yeast. eLife 2020, 9, e61245. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhao, H.; Yin, Y.; Liang, C.; Mao, X.; Liu, Y.; Yu, Q.; Li, M. Function of Atg11 in non-selective autophagy and selective autophagy of Candida albicans. Bioch. Biophys. Res. Commun. 2019, 516, 1152–1158. [Google Scholar] [CrossRef]
- Mao, X.; Yang, L.; Fan, Y.; Wang, J.; Cui, D.; Yu, D.; Yu, Q.; Li, M. The Vacuole and Mitochondria Patch(vCLAMP) Protein Mcp1 Is Involved in Maintenance of Mitochondrial Function and Mitophagy in Candida albicans. Front. Biol. 2021, 12, 633380. [Google Scholar] [CrossRef]
- Dunn, W.A., Jr.; Cregg, J.M.; Kiel, J.A.; van der Klei, I.J.; Oku, M.; Sakai, Y.; Sibirny, A.A.; Stasyk, O.V.; Veenhuis, M. Pexophagy: The selective autophagy of peroxisomes. Autophagy 2005, 1, 75–83. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Deng, Y.Z.; Naqvi, N.I. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy 2013, 9, 1818–1827. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.Z.; Qu, Z.; He, Y.; Naqvi, N.I. Sorting nexin Snx41 is essential for conidiation and mediates glutathione-based antioxidant defense during invasive growth in Magnaporthe oryzae. Autophagy 2012, 8, 1058–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Qu, Z.; Naqvi, N.I. The role of snx41-based pexophagy in magnaporthe development. PLoS ONE 2013, 8, e79128. [Google Scholar] [CrossRef] [PubMed]
- van Weering, J.R.; Verkade, P.; Cullen, P.J. SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic 2012, 13, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Yamano, K.; Sato, M.; Matsuda, N.; Okamoto, K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021, 40, e104705. [Google Scholar] [CrossRef]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.O.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, C.; Leao, M.; Soares, J.; Bessa, C.; Saraiva, L. New therapeutic strategies for cancer and neurodegeneration emerging from yeast cell-based systems. Curr. Pharm. Des. 2012, 18, 4223–4235. [Google Scholar] [CrossRef] [PubMed]
- Outeiro, T.F.; Lindquist, S. Yeast cells provide insight into alpha-synuclein biology and pathology. Science 2003, 302, 1772–1775. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.; Brandis, K.A.; Herrera, S.K.; Johnson, B.E.; Vaidya, T.; Shrestha, R.; Debburman, S.K. Alpha-synuclein budding yeast model: Toxicity enhanced by impaired proteasome and oxidative stress. J. Mol. Neurosci. 2006, 28, 161–178. [Google Scholar] [CrossRef]
- Soper, J.H.; Roy, S.; Stieber, A.; Lee, E.; Wilson, R.B.; Trojanowski, J.Q.; Burd, C.G.; Lee, V.M. α-synuclein-induced aggregation of cytoplasmic vesicles in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008, 19, 1093–1103. [Google Scholar] [CrossRef] [Green Version]
- Büttner, S.; Bitto, A.; Ring, J.; Augsten, M.; Zabrocki, P.; Eisenberg, T.; Jungwirth, H.; Hutter, S.; Carmona-Gutierrez, D.; Kroemer, G.; et al. Functional mitochondria are required for alpha-synuclein toxicity in aging yeast. J. Biol. Chem. 2008, 283, 7554–7560. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Costa, V.; Martins, L.M.; Saraiva, L. A yeast model of the Parkinson’s disease-associated protein Parkin. Exp. Cell Res. 2015, 333, 73–79. [Google Scholar] [CrossRef]
- Santos, M.M.S.; Elsztein, C.; De Souza, R.B.; Paiva, S.S.L., Jr.; de Azevêdo Silva, J.; Crovella, S.; De Morais, M.A., Jr. Respiratory deficiency in yeast mevalonate kinase deficiency may explain MKD-associate metabolic disorder in humans. Curr. Genet. 2018, 64, 871–881. [Google Scholar] [CrossRef]
- Santos, M.M.S.; Gatica, D.; de Azêvedo Silva, J.; Crovella, S.; Klionsky, D.J.; De Morais, M.A., Jr. Incomplete mitophagy in the mevalonate kinase-deficient Saccharomyces cerevisiae and its relation to the MKD-related autoinflammatory disease in humans. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166053. [Google Scholar] [CrossRef] [PubMed]
- Innokentev, A.; Furukawa, K.; Fukuda, T.; Saigusa, T.; Inoue, K.; Yamashita, S.I.; Kanki, T. Association and dissociation between the mitochondrial Far complex and Atg32 regulate mitophagy. eLife 2020, 9, e63694. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012, 14, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Tian, W.; Hu, Z.; Chen, G.; Huang, L.; Li, W.; Zhang, X.; Xue, P.; Zhou, C.; Liu, L.; et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014, 15, 566–575. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Liu, L.; Cheng, Q.; Li, Y.; Wu, H.; Zhang, W.; Wang, Y.; Sehgal, S.A.; Siraj, S.; Wang, X.; et al. Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. EMBO Rep. 2017, 18, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Han, Z.; Feng, D.; Chen, Y.; Chen, L.; Wu, H.; Huang, L.; Zhou, C.; Cai, X.; Fu, C.; et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 2014, 54, 362–377. [Google Scholar] [CrossRef] [Green Version]
- Sulistijo, E.S.; MacKenzie, K.R. Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix-helix interactions. J. Mol. Biol. 2006, 364, 974–990. [Google Scholar] [CrossRef]
- Russ, W.P.; Engelman, D.M. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 2000, 296, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Marinković, M.; Šprung, M.; Novak, I. Dimerization of mitophagy receptor BNIP3L/NIX is essential for recruitment of autophagic machinery. Autophagy 2021, 17, 1232–1243. [Google Scholar] [CrossRef]
- Böckler, S.; Westermann, B. Mitochondrial ER Contacts Are Crucial for Mitophagy in Yeast. Dev. Cell. 2014, 28, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Belgareh-Touzé, N.; Cavellini, L.; Cohen, M.M. Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy. Autophagy 2017, 13, 114–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onishi, M.; Nagumo, S.; Iwashita, S.; Okamoto, K. The ER membrane insertase Get1/2 is required for efficient mitophagy in yeast. Biochem. Biophys. Res. Commun. 2018, 503, 14–20. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatia-Kissova, I.; Camougrand, N. Mitophagy in Yeast: Decades of Research. Cells 2021, 10, 3541. https://doi.org/10.3390/cells10123541
Bhatia-Kissova I, Camougrand N. Mitophagy in Yeast: Decades of Research. Cells. 2021; 10(12):3541. https://doi.org/10.3390/cells10123541
Chicago/Turabian StyleBhatia-Kissova, Ingrid, and Nadine Camougrand. 2021. "Mitophagy in Yeast: Decades of Research" Cells 10, no. 12: 3541. https://doi.org/10.3390/cells10123541
APA StyleBhatia-Kissova, I., & Camougrand, N. (2021). Mitophagy in Yeast: Decades of Research. Cells, 10(12), 3541. https://doi.org/10.3390/cells10123541





