Evodiamine Stabilizes Topoisomerase I-DNA Cleavable Complex to Inhibit Topoisomerase I Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Growth inhibition of EVO
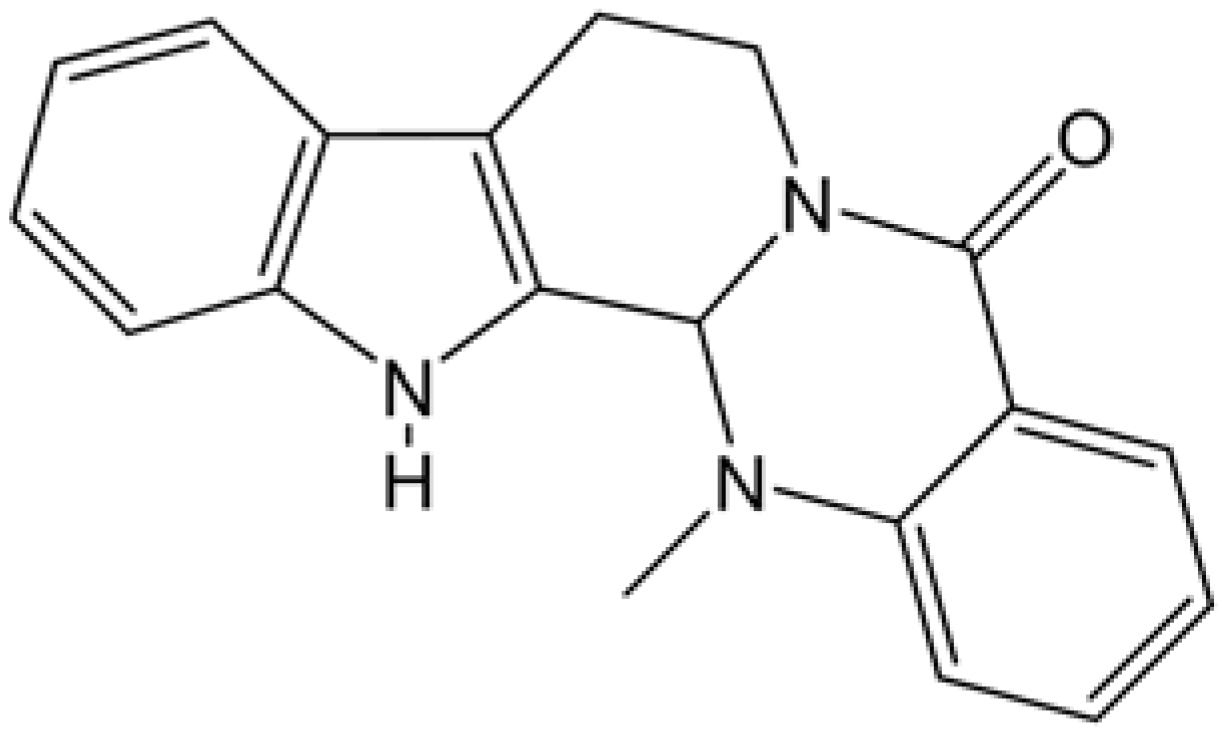
2.2. Inhibition of supercoiled DNA relaxation from Top I catalysis by EVO

2.3. EVO depletes the Top I protein

2.4. TopI-DNA complex-trapping activity in MCF-7 cells
2.5. Proposed TopI-inhibition mechanism of EVO


3. Experimental
3.1 Materials
3.2 Cell culture
3.3. Preparation of Nuclear Extract
3.4. Recombinant human TopI (hTopI) protein expression and purification
3.5. TopI-catalyzed supercoiled DNA relaxation
3.6. KCl/SDS precipitation assay of the covalent TopI-DNA complex
3.7. Detection of the protein levels of TopI
3.8. MTT cell viability assay
4. Conclusions
Acknowledgements
References and Notes
- Teicher, B.A. Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem. Pharmacol. 2008, 75, 1262–1271. [Google Scholar] [CrossRef]
- Liu, L.F. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 1989, 58, 351–375. [Google Scholar] [CrossRef]
- Tsao, Y.P.; D'Arpa, P.; Liu, L.F. The involvement of active DNA synthesis in camptothecin-induced G2 arrest: altered regulation of p34cdc2/cyclin B. Cancer Res. 1992, 52, 1823–1829. [Google Scholar]
- Niina, I.; Uchiumi, T.; Izumi, H.; Torigoe, T.; Wakasugi, T.; Igarashi, T.; Miyamoto, N.; Onitsuka, T.; Shiota, M.; Okayasu, R.; Chijiiwa, K.; Kohno, K. DNA topoisomerase inhibitor, etoposide, enhances GC-box-dependent promoter activity via Sp1 phosphorylation. Cancer Sci. 2007, 98, 858–863. [Google Scholar] [CrossRef]
- Feun, L.; Savaraj, N. Topoisomerase I inhibitors for the treatment of brain tumors. Expert Rev. Anticancer Ther. 2008, 8, 707–716. [Google Scholar] [CrossRef]
- Wethington, S.L.; Wright, J.D.; Herzog, T.J. Key role of topoisomerase I inhibitors in the treatment of recurrent and refractory epithelial ovarian carcinoma. Expert Rev. Anticancer Ther. 2008, 8, 819–831. [Google Scholar] [CrossRef]
- Sadaie, M.R.; Mayner, R.; Doniger, J. A novel approach to develop anti-HIV drugs: adapting non-nucleoside anticancer chemotherapeutics. Antiviral Res. 2004, 61, 1–18. [Google Scholar] [CrossRef]
- Anderson, V.E.; Osheroff, N. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr. Pharm. Des. 2001, 7, 337–353. [Google Scholar] [CrossRef]
- Song, J.; Parker, L.; Hormozi, L.; Tanouye, M.A. DNA topoisomerase I inhibitors ameliorate seizure-like behaviors and paralysis in a Drosophila model of epilepsy. Neuroscience 2008, 156, 722–728. [Google Scholar] [CrossRef]
- Verdrengh, M.; Tarkowski, A. Impact of topoisomerase II inhibition on cytokine and chemokine production. Inflamm. Res. 2003, 52, 148–153. [Google Scholar] [CrossRef]
- Wang, T.; Wang, Y.; Kontani, Y.; Kobayashi, Y.; Sato, Y.; Mori, N.; Yamashita, H. Evodiamine improves diet-induced obesity in a uncoupling protein-1-independent manner: involvement of antiadipogenic mechanism and extracellularly regulated kinase/mitogen-activated protein kinase signaling. Endocrinology 2008, 149, 358–366. [Google Scholar] [CrossRef]
- Ogasawara, M.; Matsunaga, T.; Takahashi, S.; Saiki, I.; Suzuki, H. Anti-invasive and metastatic activities of evodiamine. Biol. Pharm. Bull. 2002, 25, 1491–1493. [Google Scholar] [CrossRef]
- Ko, H.C.; Wang, Y.H.; Liou, K.T.; Chen, C.M.; Chen, C.H.; Wang, W.Y.; Chang, S.; Hou, Y.C.; Chen, K.T.; Chen, C.F.; Shen, Y.C. Anti-inflammatory effects and mechanisms of the ethanol extract of Evodia rutaecarpa and its bioactive components on neutrophils and microglial cells. Eur. J. Pharmacol. 2007, 555, 211–217. [Google Scholar] [CrossRef]
- Kan, S.F.; Huang, W.J.; Lin, L.C.; Wang, P.S. Inhibitory effects of evodiamine on the growth of human prostate cancer cell line LNCaP. Int. J. Cancer. 2004, 110, 641–651. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, S.H.; Jiang, R.W.; Cai, M.; Liu, X.; Ding, L.S.; Xu, H.X.; But, P.P.; Shaw, P.C. Quantitative analyses of indoloquinazoline alkaloids in Fructus Evodiae by high-performance liquid chromatography with atmospheric pressure chemical ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 3111–3118. [Google Scholar] [CrossRef]
- Shuman, S.; Golder, M.; Moss, B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J. Biol. Chem. 1988, 263, 16401–16407. [Google Scholar]
- Gupta, M.; Zhu, C.X.; Tse-Dinh, Y.C. An engineered mutant of vaccinia virus DNA topoisomerase I is sensitive to the anti-cancer drug camptothecin. J. Biol. Chem. 1992, 267, 24177–24180. [Google Scholar]
- Chu, X.Y.; Kato, Y.; Sugiyama, Y. Multiplicity of biliary excretion mechanisms for irinotecan, CPT-11, and its metabolites in rats. Cancer Res. 1997, 57, 1934–1938. [Google Scholar]
- Yang, C.H.; Schneider, E.; Kuo, M.L.; Volk, E.L.; Rocchi, E.; Chen, Y.C. BCRP/MXR/ABCP expression in topotecan-resistant human breast carcinoma cells. Biochem. Pharmacol. 2000, 60, 831–837. [Google Scholar] [CrossRef]
- Xu, M.L.; Li, G.; Moon, D.C.; Lee, C.S.; Woo, M.H.; Lee, E.S.; Jahng, Y.; Chang, H.W.; Lee, S.H.; Son, J.K. Cytotoxicity and DNA topoisomerase inhibitory activity of constituents isolated from the fruits of Evodia officinalis. Arch. Pharm Res. 2006, 29, 541–547. [Google Scholar] [CrossRef]
- Liao, C.H.; Pan, S.L.; Guh, J.H.; Chang, Y.L.; Pai, H.C.; Lin, C.H.; Teng, C.M. Antitumor mechanism of evodiamine, a constituent from Chinese herb Evodiae fructus, in human multiple-drug resistant breast cancer NCI/ADR-RES cells in vitro and in vivo. Carcinogenesis 2005, 26, 968–975. [Google Scholar]
- Zhelkovsky, A.M.; Moore, C.L. Overexpression of human DNA topoisomerase I in insect cells using a baculovirus vector. Protein Expr. Purif. 1994, 5, 364–370. [Google Scholar] [CrossRef]
- Yang, M.; Hsu, C.T.; Ting, C.Y.; Liu, L.F.; Hwang, J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J. Biol. Chem. 2006, 281, 8264–8274. [Google Scholar]
- Sekiguchi, J.; Cheng, C.; Shuman, S. Kinetic analysis of DNA and RNA strand transfer reactions catalyzed by vaccinia topoisomerase. J. Biol. Chem. 1997, 272, 15721–15728. [Google Scholar] [CrossRef]
- Ting, C.Y.; Hsu, C.T.; Hsu, H.T.; Su, J.S.; Chen, T.Y.; Tarn, W.Y.; Kuo, Y.H.; Whang-Peng, J.; Liu, L.F.; Hwang, J. Isodiospyrin as a novel human DNA topoisomerase I inhibitor. Biochem. Pharmacol. 2003, 66, 1981–1991. [Google Scholar] [CrossRef]
- Yoshinari, T.; Yamada, A.; Uemura, D.; Nomura, K.; Arakawa, H.; Kojiri, K.; Yoshida, E.; Suda, H.; Okura, A. Induction of topoisomerase I-mediated DNA cleavage by a new indolocarbazole, ED-110. Cancer Res. 1993, 53, 490–494. [Google Scholar]
- Hwong, C.L.; Chen, C.Y.; Shang, H.F.; Hwang, J. Increased synthesis and degradation of DNA topoisomerase I during the initial phase of human T lymphocyte proliferation. J. Biol. Chem. 1993, 268, 18982–18986. [Google Scholar]
- Lin, H.C.; Tsai, S.H.; Chen, C.S.; Chang, Y.C.; Lee, C.M.; Lai, Z.Y.; Lin, C.M. Structure-activity relationship of coumarin derivatives on xanthine oxidase-inhibiting and free radical-scavenging activities. Biochem. Pharmacol. 2008, 75, 1416–1425. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chan, A.L.-F.; Chang, W.-S.; Chen, L.-M.; Lee, C.-M.; Chen, C.-E.; Lin, C.-M.; Hwang, J.-L. Evodiamine Stabilizes Topoisomerase I-DNA Cleavable Complex to Inhibit Topoisomerase I Activity. Molecules 2009, 14, 1342-1352. https://doi.org/10.3390/molecules14041342
Chan AL-F, Chang W-S, Chen L-M, Lee C-M, Chen C-E, Lin C-M, Hwang J-L. Evodiamine Stabilizes Topoisomerase I-DNA Cleavable Complex to Inhibit Topoisomerase I Activity. Molecules. 2009; 14(4):1342-1352. https://doi.org/10.3390/molecules14041342
Chicago/Turabian StyleChan, Agnes L.-F., Wen-Shin Chang, Li-Min Chen, Chi-Ming Lee, Chiao-En Chen, Chun-Mao Lin, and Jau-Lang Hwang. 2009. "Evodiamine Stabilizes Topoisomerase I-DNA Cleavable Complex to Inhibit Topoisomerase I Activity" Molecules 14, no. 4: 1342-1352. https://doi.org/10.3390/molecules14041342
APA StyleChan, A. L.-F., Chang, W.-S., Chen, L.-M., Lee, C.-M., Chen, C.-E., Lin, C.-M., & Hwang, J.-L. (2009). Evodiamine Stabilizes Topoisomerase I-DNA Cleavable Complex to Inhibit Topoisomerase I Activity. Molecules, 14(4), 1342-1352. https://doi.org/10.3390/molecules14041342




