Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets?
Abstract
:1. Introduction
2. General Structures of Vitamin K
3. Vitamin K1 in Humans


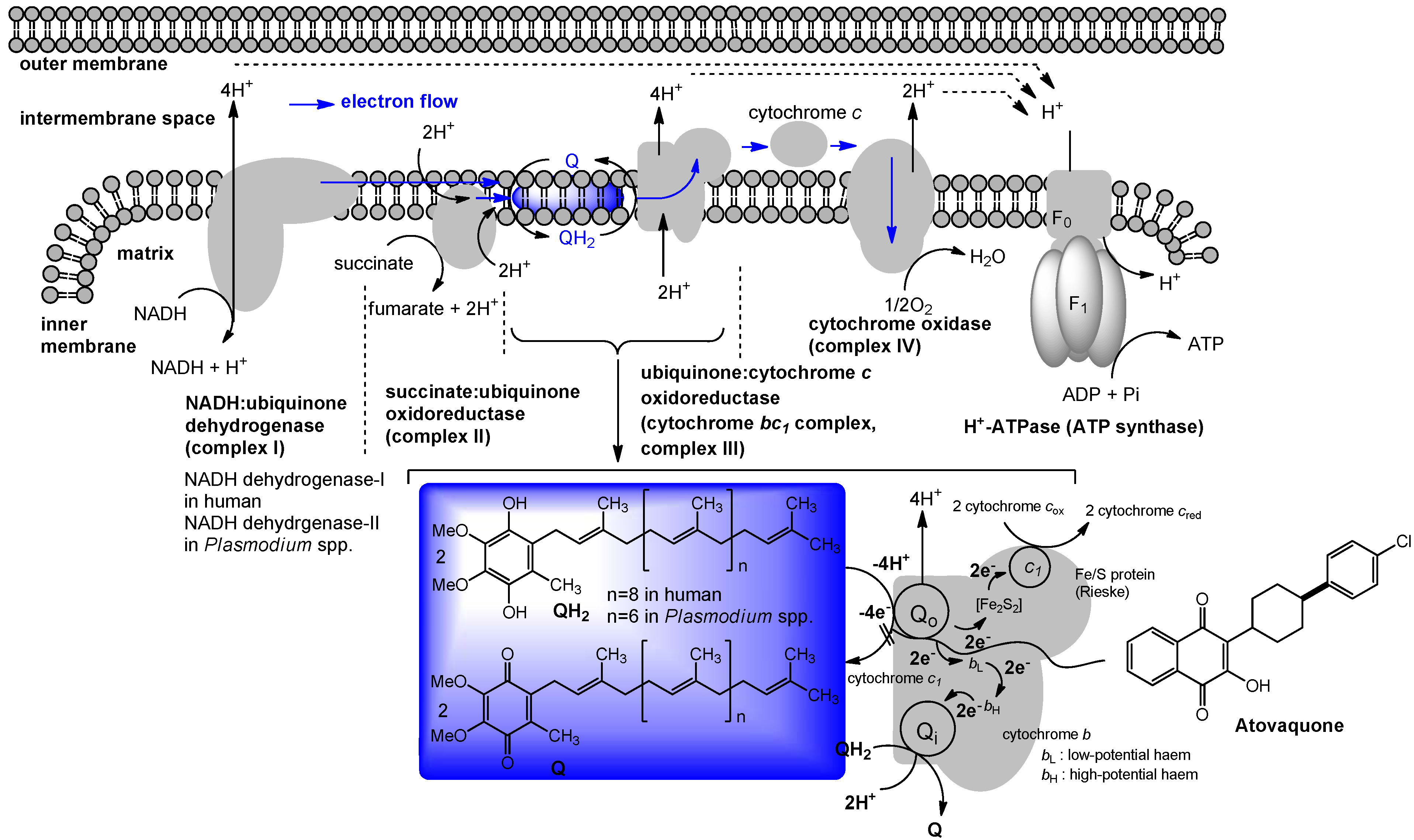
4. Ubiquinone in Humans
5. The Role of Vitamin K2 in Electron Transport
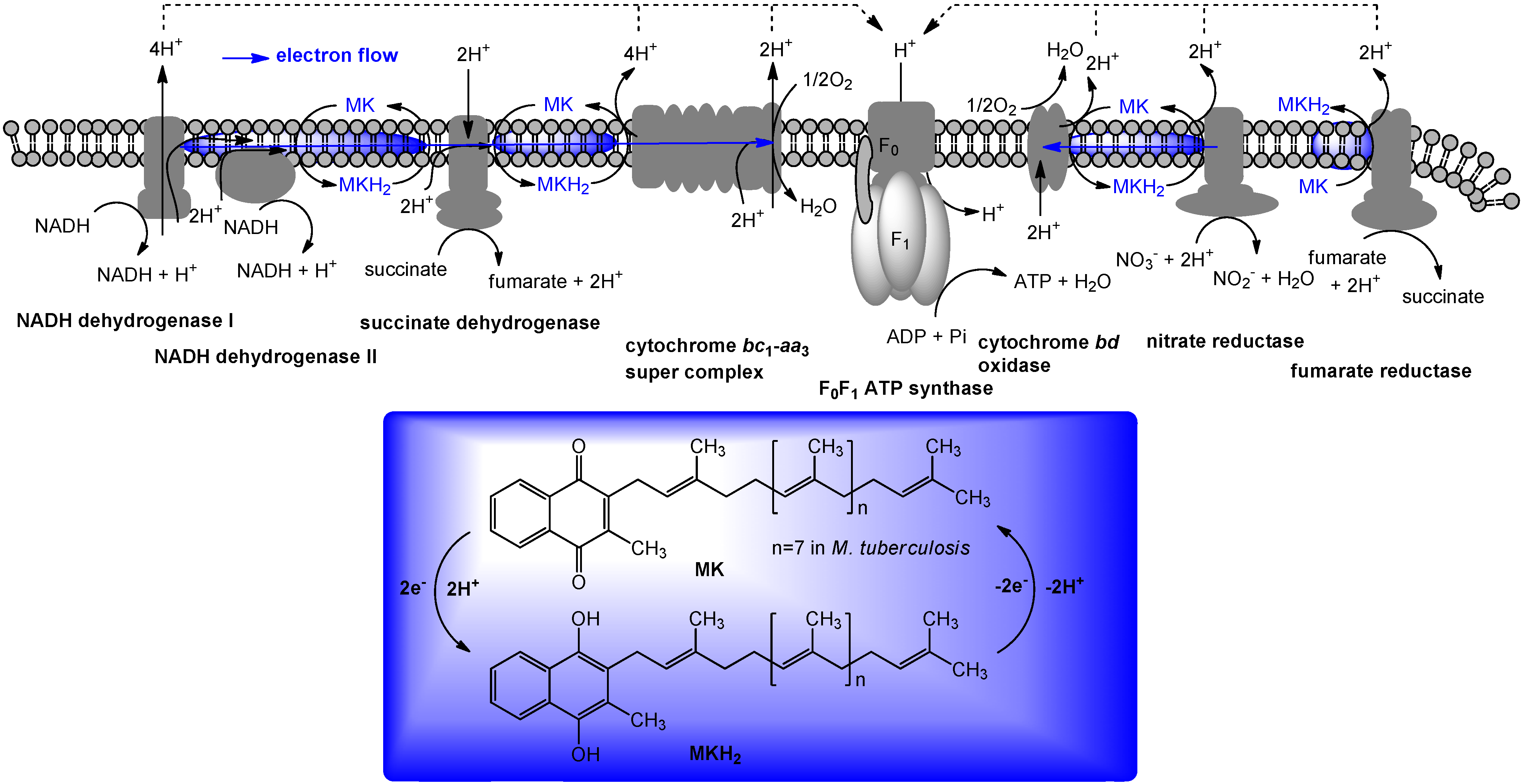
6. Biosynthesis of Menaquinone

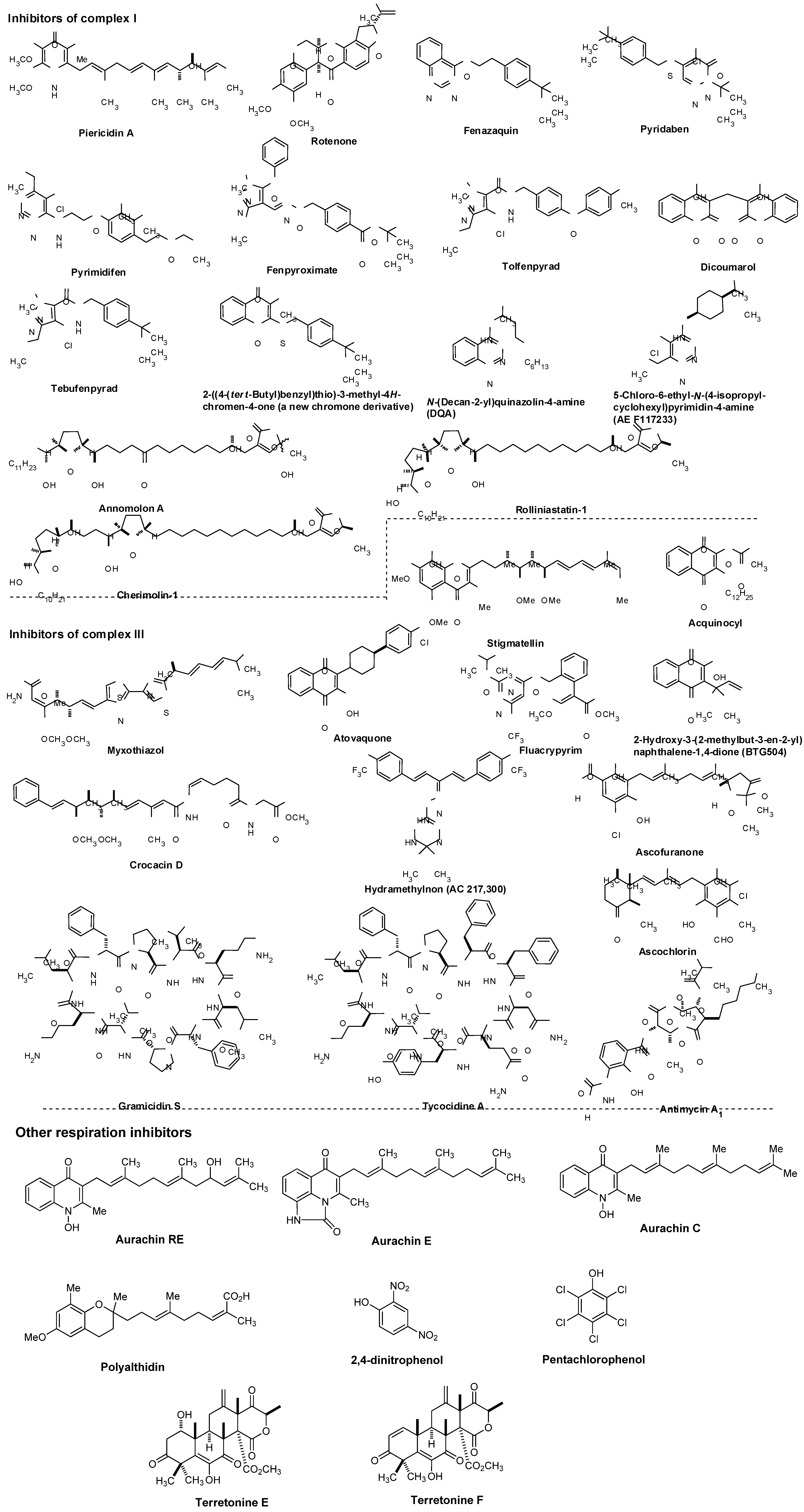
7. Targeting Electron Transport System in Drug Discovery
7.1. NADH Dehydrogenase

7.2. Other Respiration Inhibitors
8. Menaquinone Biosynthesis as a Target for Antibacterial Agents

9. Summary
Acknowledgements
References and Notes
- Usui, Y.; Tanimura, H.; Nishimura, N.; Kobayashi, N. Vitamin K concentrations in the plasma and liver of surgical patients. Am. J. Clin. Nutr. 1990, 51, 846–852. [Google Scholar]
- Kindberg, C.G.; Suttie, J.W. J. W. Effect of various intakes of phylloquinone on signs of vitamin K deficiency and serum and liver phylloquinone concentrations in the rat. Vitamins 1989, 175–180. [Google Scholar]
- Collins, M.D.; Jones, D. Distribution of isoprenoid quinone structual types in bacterial and their taxonomic implications. Microbiol. Rev. 1981, 45, 316–354. [Google Scholar]
- Nomenclature of quinones with isoprenoid side-chains. Pure Appl. Chem. 1974, 38, 439–447. [CrossRef]
- Suttie, J.W. Synthesis of vitamin K dependent proteins. FASEB 1993, 7, 445–452. [Google Scholar]
- Cain, D.; Hutson, S.M.; Wallin, R. Assembly of the warfarin-sensitive vitamin K 2,3-epoxide reductase. J. Biol. Chem. 1997, 272, 29068–29075. [Google Scholar]
- Jin, D-Y.; Tie, J-K.; Stafford, D.W. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K. Biochemistry 2007, 46, 7279–7283. [Google Scholar] [CrossRef]
- Vogel, H.J.; Brokx, R.D.; Ouyang, H. Calcium-binding proteins. Method. Mol. Biol. 2002, 172, 1940–6029. [Google Scholar]
- Francis, C.W. Warfarin: An historical perspective. Hematology 2008, 251. [Google Scholar] [CrossRef]
- Gebauer, M. Synthesis and structure-activity relationships of novel warfarin derivatives. Bioorg. Med. Chem. 2007, 15, 2414–2420. [Google Scholar] [CrossRef]
- Kuruvilla, M.; Gurk-Turner, C. A review of warfarin dosing and monitoring. BUMC Proc. 2001, 14, 305–306. [Google Scholar]
- Vermeer, C.; Gijsbers, B.L.; Craciun, A.M.; Groenen-van Dooren, M.M.C.L.; Knapen, M.H. Effects of vitamin K on bone mass and bone metabolism. Am. Inst. Nutr. 1996, 1187S–1191S. [Google Scholar]
- Iwamoto, J.; Yeh, J.K.; Takeda, T.; Ichimura, S.; Sato, Y. Comparative effects of vitamin K and vitamin D supplementation on prevention of osteopenia in calcium-deficient young rats. Bone 2003, 33, 557–566. [Google Scholar] [CrossRef]
- Visser, C.M. Some speculations on the mechanisms of the vitamins E and K starting from origin of life considerations and the antioxidant theory. Bioorg. Chem. 1980, 9, 411–422. [Google Scholar] [CrossRef]
- Shirakawa, H.; Ohsaki, Y.; Minegishi, Y.; Takumi, N.; Ohinata, K.; Furukawa, Y.; Mizutani, T.; Komai, M. Vitamin K deficiency reduces testosterone production in the testis through down-regulation of the Cyp11a a cholesterol side chain cleavage enzyme in rats. Biochim. Biophys. Acta 1760, 1482–1488. [Google Scholar]
- Awato, K. Prostate and menadiol sodium diphosphate. 1. Menadiol sodium diphosphate as a new substrate for measuring acid phosphatase activity and a discussion of prostatic tumor models. Nippon Hinyokika Gakkai Zasshi 1982, 73, 507–515. [Google Scholar]
- Munter, G.; Hershko, C. Increased warfarin sensitivity as an early manifestation of occult prostate cancer with chronic disseminated intravascular coagulation. Act. Haematol. 2001, 105, 97–99. [Google Scholar] [CrossRef]
- Nakamura, M., Nagano; Noda, T.; Wada, H.; Ota, H.; Damdinsuren, B.; Marubashi, S.; Miyamoto, A.; Takeda, Y.; Doki, Y.; Umeshita, K.; Dono, K.; Sakon, M.; Monden, M. Vitamin K2 has growth inhibition effect against hepatocellular carcinoma cell lines but does not enhance anti-tumor effect of combination treatment of interferon- and fluorouracil in vitro. Hepatol. Res. 2006, 35, 289–295. [Google Scholar]
- Thijssen, H.H.W.; Drittij-Reijnders, M.J. Vitamin K status in human tissues: tissue-specific accumulation of phylloquinone and menaquinone-4. British J. Nutr. 1996, 75, 121–127. [Google Scholar] [CrossRef]
- Yoshitomo, S.; Shinya, A.; Aya, M.; Yuka, S.; Kimie, N.; Maya, K.; Naoko, T.; Toshio, O. Synthesis and development of biologically active fluorescent-labeled vitamin K analogues and monitoring of their subcellular distribution. Tetrahedron 2008, 64, 8789–8796. [Google Scholar]
- Toshio, O.; Kimie, N.; Maya, K. Vitamin K and bone update. In vivo metabolism of vitamin K. - in relation to the conversion of vitamin K1 to MK-4. Clin. Calcium 2009, 19, 1779–1787. [Google Scholar]
- Usui, Y.; Tanimura, H.; Nishimura, N.; Kobayashi, N.; Okanoue, T.; Ozawa, K. Vitamin K concentrations in the plasma and liver of surgical patients. Am. J. Clin. Nutr. 1990, 5, 846–852. [Google Scholar]
- Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef]
- Mitchell, P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. Cambridge Phil Soc. 1966, 41, 445–502. [Google Scholar] [CrossRef]
- Mitchell, P. Chemiosmotic coupling in energy transduction: A logical development of biochemical knowledge. Bioenergetics 1972, 3, 5–24. [Google Scholar] [CrossRef]
- Mellors, A.; Tappel, A.L. The inhibition of mitochondrial peroxidation by ubiquinone and ubiquinol. J. Biol. Chem. 1966, 241, 4353–4356. [Google Scholar]
- Linnane, A.W.; Zhang, C.; Yarovaya, N.; Kopsidas, G.; Kovalenko, S.; Papakostopoulos, P.; Eastwood, H.; Gravens, S.; Richardson, M. Human aging and global function of Coenzyme Q10. Ann. N.Y. Acad. Sci. 2002, 959, 396–411. [Google Scholar]
- Harman, D. Free radicals in aging. Mol. Cell. Biochem. 1988, 84, 55–61. [Google Scholar]
- Emster, L.; Dallner, G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim. Biophys. Acta 1995, 1271, 195–204. [Google Scholar] [CrossRef]
- Folkers, K.; Langsjoen, P.H.; Willis, R.; Richardson, P.; Xia, L.; Ye, C.; Tamagawa, H. (1990) Lovastatin decreases coenzyme Q levels in humans. Proc. Natl. Acad Sci. 1990, 87, 8931–8934. [Google Scholar]
- Szkopisńka, A. Ubiquinone. Biosynthesis of quinone ring and its isoprenoid side chain. Intracellular localization. Acta Biochim. Pol. 2000, 47, 469–480. [Google Scholar]
- Haddock, B.A.; Colin, J.W. Bacterial respiration. Bacteriol. Rev. 1977, 41, 47–99. [Google Scholar]
- Yasuhiro, A. Bacterial electron transport chains. Ann. Rev. Biochem. 1988, 57, 101–312. [Google Scholar] [CrossRef]
- Suvana, K.D.; Stevenson, R.; Meganathan, R.; Hudspeth, M.E.S. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J. Bacteriol. 1998, 180, 2782–2787. [Google Scholar]
- Truglio, J.J.; Theis, K.; Feng, Y.; Gajda, R.; Machutta, C.; Tong, P.J.; Kisher, C. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J. Biol. Chem. 2003, 24, 42352–4236. [Google Scholar]
- Bishop, D.H.L.; Pandya, K.P.; King, H.K. Ubiquinone and vitamin K in bacteria. Biochem. J. 1962, 83, 606–614. [Google Scholar]
- Das, A.; Hugenholtz, J.; van Halbeek, H.; Ljungdal, L.G. Structure and function of a menaquinone involved in electron transport in membranes of Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 1989, 171, 5823–5829. [Google Scholar]
- Martin, J.L.; M. McMillan, F. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar]
- Bentley, R.; Meganathan, R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 1982, 46, 241–280. [Google Scholar]
- Bentley, R. Biosynthesis of vitamin K and other natural naphthoquinones. Pure Appl. Chem. 1975, 41, 47–68. [Google Scholar] [CrossRef]
- Shineberg, B.; Young, I.G. Biosynthesis of bacterial menaquinones: the membrane-associated 1,4-dihydroxy-2-naphthoate octaprenyltransferase of Escherichia coli. Biochemistry 1976, 15, 2754–2758. [Google Scholar] [CrossRef]
- Meganathan, R.; Bentley, R. Biosynthesis of o-succinylbenzoic acid in a men-Escherichia coli mutant requires decarboxylation of L-glutamate at the C-1 position. Biochemistry 1981, 20, 5336–5340. [Google Scholar] [CrossRef]
- Widhalm, J.R.; van Oostende, C.; Furt, F.; Basset, GJC. A dedicated thioesterase of the Hotdog-fold family is required for the biosynthesis of the naphthoquinone ring of vitamin K1. Proc. Natl. Acad. Sci. USA 2009, 106, 5599–5603. [Google Scholar]
- Schoepp-Cotheneta, B.; Lieutauda, C.; Baymanna, F.; Vermégliob, A.; Friedrichc, T.; Kramer, D.M.; Nitschke, W. Menaquinone as pool quinone in a purple bacterium. Proc. Natl. Acad. Sci. USA 2009, 106, 8549–8554. [Google Scholar]
- Hiratsuka, T.; Furihata, K.; Ishikawa, J.; Yamashita, H.; Itoh, N.; Seto, H.; Dair, T. An Alternative menaquinone biosynthetic pathway operating in microorganisms. Science 2008, 321, 1670–1673. [Google Scholar] [CrossRef]
- Shaw, D.J.; Rice, D.W.; Guest, J.R. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J. Mol. Biol. 1983, 166, 241–247. [Google Scholar] [CrossRef]
- Unden, G. Differential roles for menaquinone and demethylmenaquinone in anaerobic electron transport of E. coli and their fnr-independent expression. Arch. Microbiol. 1988, 150, 499–503. [Google Scholar] [CrossRef]
- Kaiser, M.; Sawers, G. Overlapping promoters modulate Fnr- and ArcA-dependent anaerobic transcriptional activation of the focApfl operon in Escherichia coli. Microbiology 1997, 143, 775–783. [Google Scholar] [CrossRef]
- Zigha, A.; Rosenfeld, E.; Schmitt, P.; Duport, C. The redox regulator Fnr is required for fermentative growth and enterotoxin synthesis in bacillus cereus F4430/73. J. Bacteriol. 2007, 189, 2813–2824, and references therein. [Google Scholar] [CrossRef]
- Hollfinder, R. The dependence on quinone specificity of terminal electron transport of bacteria. Curr. Microbiol. 1981, 6, 155–159. [Google Scholar]
- Carlone, G.M.; Schalla, W.O.; Moss, C. W.; Ashlley, D.L.; Fast, D.M.; Holler, J.S.; Plikaytis, B.D. Haemophilus ducreyi isoprenoid quinone content and structure determination. Int. J. Syst. Bacteriol. 1988, 38, 249–253. [Google Scholar] [CrossRef]
- Kröger, A.; Unden, G. The function of menaquinone in bacterial electron transpor. In Coenzyme Q; Lenz, G., Ed.; John Wiley & Sons: New York, NY, USA, 1985; pp. 285–300. [Google Scholar]
- Vaidya, A.B.; Paintera, H.J.; Morriseya, J.M.; Mathera, M.W. The validity of mitochondrial dehydrogenases as antimalarial drug targets. Trends Parasitol. 2007, 24, 8–9. [Google Scholar]
- Painter, H.J.; Morrisey, J.M.; Mather, M.W.; Vaidya, A.B. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature 2007, 446, 88–91. [Google Scholar]
- Smilkstein, M.J.; Forquer, I.; Kanazawa, A.; Kelly, J.X.; Rolf, W.; Winter, R.W.; Hinrichs, D.J.; Kramer, D.M.; Riscoe, M.K. Mol. Biochem. Parasitol. 2008, 159, 64–68. [CrossRef]
- Kessl, J.J.; Lange, B.B.; Mertbitz-Zahradnik, T.; Hill, P.; Meunier, B.; Palsdottir, H.; Hunte, C.; Meshnilk, S.; Trumpower, B.L. Molecular basis of atovaquone binding to the cytochrome bc1 complex. J. Biol. Chem. 2003, 278, 31312–31318. [Google Scholar]
- Weistein, E.A.; Yano, T.; Li, L-S.; Avarbock, D.; Avarbock, A.; Helm, D.; McColm, A.A.; Duncan, K.; Lonsdale, J.T.; Rubin, H. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. USA 2005, 102, 4548–4553. [Google Scholar]
- Shi, L.; Sohaskey, C.D.; Kana, B.D.; Dawes, S.; North, R.J.; Mizrahi, V.; Gennaro, M.L. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. USA 2005, 102, 15629–15634. [Google Scholar]
- Yano, T.; Li, L.S.; Weinstein, E.; Teh, J.S.; Rubin, H. Steady-state kinetics and inhibitory action of antitubercular phenothiazines on Mycobacterium tuberculosis Type-II NADH-menaquinone oxidoreductase (NDH-2). J. Biol. Chem. 2006, 281, 11456–11463. [Google Scholar]
- Dekeyser, M. Acaricide mode of action. Pest Manag. Sci. 2005, 61, 103–110. [Google Scholar] [CrossRef]
- Estornell, E. Mitochondrial complex I: new insights from inhibitor assays. Plotoplasma 2000, 213, 11–17. [Google Scholar] [CrossRef]
- Sharova, I.V.; Vekshin, N.L. Rotenone-insensitive NADH oxidation in mitochondrial suspension is performed by NADH dehydrogenase of respiratory chain fragments. Biofizika 2004, 49, 814–821. [Google Scholar]
- Bai, Y.L. Acaricides with different modes of action. Xiandai Nongyao 2005, 4, 27–30. [Google Scholar]
- Wood, E.; Latli, B.; Casida, J.E. Fenazaquin acaricide specific binding sites in NADH:ubiquinone oxidoreductase and apparently the ATP synthase stalk. Pestic. Biochem. Physiol. 1996, 54, 135–145. [Google Scholar] [CrossRef]
- Hochstein, L.I.; Cronin, S.E. Electron transport in Paracoccus halodenitrificans and the role of ubiquinone. NASA Tech. Memo. 1983, 23, 572–577. [Google Scholar]
- Lindell, S.D.; Ort, O.; Lümmen, P.; Klein, R. The design and synthesis of novel inhibitors of NADH:ubiquinone oxidoreductase. Bioorg. Med. Chem. Lett. 2004, 14, 511–514. [Google Scholar] [CrossRef]
- Beautement, K.; Clough, J.M.; de Fraine, P.J.; Godfrey, C.R.A. Fungicidal beta-methoxyacrylates: from natural products to novel synthetic agricultural fungicides. Pestic Sci. 1991, 31, 499–519. [Google Scholar] [CrossRef]
- Bermejo, A.; Figadere, B.; Zafra-Polo, M-C.; Barrachina, I.; Estornell, E.; Cortes, D. Acetogenins from Annonaceae: Recent progress in isolation, synthesis and mechanisms of action. Nat. Prod. Rep. 2005, 22, 269–303. [Google Scholar] [CrossRef]
- Son, J.K.; Kim, D. H.; Woo, M.H. Two new epimeric pairs of acetogenins bearing a carbonyl group from Annona cherimolia seeds. J. Nat. Prod. 2003, 66, 1369–1372. [Google Scholar] [CrossRef]
- Crofts, A.R. the cytochrome bc1 complex: Function in the context of structure. Annu. Rev. Physiol. 2004, 66, 689–733. [Google Scholar] [CrossRef]
- Xia, D.; Yu, C.A.; Kim, H.; Xia, J.Z.; Kachurin, A.M.; Zhang, L.; Yu, L.; Deisenhofer, J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 1997, 277, 60–66. [Google Scholar]
- Smith, J.L.; Zhang, H.; Yan, J.; Kurisu, G.; Cramer, W.A. Cytochrome bc complexes: a common core of structure and function surrounded by diversity in the outlying provinces. Curr. Opini. Struct. Biol. 2004, 14, 432–439. [Google Scholar] [CrossRef]
- Iwata, S.; Lee, J.W.; Okada, K.; Lee, J.K.; Iwata, M.; Ramussen, B.; Link, T.A.; Ramaswamy, S.; Jap, B.K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 1998, 281, 64–71. [Google Scholar]
- Hunte, C.; Koepke, J.; Lange, C.; Rossmanith, T.; Michel, H. Structure at 2.3 A resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure (Lond). 2000, 8, 669–684. [Google Scholar]
- Zhang, Z.; Huang, L.; Shulmeister, V.M.; Chi, Y.I.; Kim, K.K.; Huang, L.W.; Crofts, A.R.; Berry, E.A.; Kim, S.H. Electron transfer by domain movement in cytochrome bc1. Nature 1998, 392, 677–684. [Google Scholar]
- Lange, C.; Hunte, C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc. Natl. Acad. Sci. USA 2002, 99, 2800–2805. [Google Scholar] [CrossRef]
- Xia, D.; Esser, L.; Elberry, M.; Zhou, F.; Yu, L.; Yu, C.A. The road to the crystal structure of the cytochrome bc1 complex from the anoxigenic, photosynthetic bacterium Rhodobacter sphaeroides. J. Bioenerg Biomembr. 2008, 40, 485–92. [Google Scholar] [CrossRef]
- Tappel, A.L. Inhibition of electron transport by antimycin A, alkyl hydroxy naphthoquinones and metal coordination compounds. Biochem. Pharmacol. 1960, 3, 289–296. [Google Scholar]
- Huang, L.S.; Cobessi, D.; Tung, E.Y.; Berry, E.A. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: A new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J. Mol. Biol. 2005, 351, 573–597. [Google Scholar] [CrossRef]
- Kotova, E.A.; Oleskin, A.V.; Samuilov, V.D. Effect of myxothiazol on the electrogenic redox chain of purple photosynthetic bacteria. Photobiochem. Photobiophys. 1983, 6, 211–221. [Google Scholar]
- Crowley, P.J.; Aspinall, I.H.; Gillen, K.; Godfrey, C.R.A.; Devillers, I.M.; Munns, G.R.; Sageot, O-A; Swanborough, J.; Worthington, P.A.; Williams, J. The crocacins: Novel natural products as leads for agricultural fungicides. Chimia 2003, 57, 685–691. [Google Scholar] [CrossRef]
- Gurung, B.; Yu, L.; Yu, C.A. Stigmatellin induces reduction of iron-sulfur protein in the oxidized cytochrome bc1 complex. J. Biol. Chem. 2008, 283, 28087–28094. [Google Scholar] [CrossRef]
- van Nieuwenhuyse, P.; van Leeuwen, T.; Khajehali, J.; Vanholme, B.; Tirry, L. Mutations in the mitochondrial cytochrome b of Tetranychus urticae Koch (Acari: Tetranychidae) confer cross-resistance between bifenazate and acequinocyl. Pest. Manag. Sci. 2009, 65, 404–412. [Google Scholar]
- Koura, Y.; Kinoshita, S.; Takasuka, K.; Koura, S.; Osaki, N.; Matsumoto, S.; Miyoshi, H. Respiratory inhibition of acaricide AKD-2023 and its deacetyl metabolite. Nihon Noyaku Gakkaishi (J. Pestic. Sci.) 1998, 23, 18–21. [Google Scholar]
- Caboni, P.; Sarais, G.; Melis, M.; Cabras, M.; Cabras, P. Determination of acequinocyl and hydroxyacequinocyl on fruits and vegetables by HPLC-DAD. J. Agric. Food Chem. 2004, 52, 6700–6702. [Google Scholar]
- Takatsuki, A; Tamura, G.; Arima, K. Antiviral and antitumor antibiotics. XIV. Effects of ascochlorin and other respiration inhibitors on multiplication of Newcastle disease virus in cultured cells. Appl. Microbiol. 1969, 17, 825–829. [Google Scholar]
- Berry, E.A.; Huang, L.S.; Lee, D.W.; Daldal, F.; Nagai, K.; Minagawa, N. Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc1 complex. Biochem. Biophy. Acta 1797, 360–370. [Google Scholar]
- Song, C.; Scharf, M.E. Mitochondrial impacts of insecticidal formate esters in insecticide-resistant and insecticide-susceptible Drosophila melanogaster. Pest. Manag. Sci. 2009, 65, 697–703. [Google Scholar] [CrossRef]
- Hong, S.; Park, K.K.; Magae, J.; Ando, K.; Lee, T.S.; Kwon, T.K.; Kwak, J.Y.; Kim, C.H.; Chang, Y.C. Ascochlorin inhibits matrix metalloproteinase-9 expression by suppressing activator protein-1-mediated gene expression through the ERK1/2 signaling pathway. J. Bio. Chem. 2005, 280, 25202–25209. [Google Scholar]
- Marques, M.A.; Citronb, D.M.; Wang, C.C. Development of tyrocidine A analogues with improved antibacterial activity. Bioorg. Med. Chem. 2007, 15, 6667–6677. [Google Scholar] [CrossRef]
- Romagnoli, S.; Oettmeier, W.; Zannoni, D. The effects of decyl aurachins C and D on the respiratory electron flow of facultative phototrophic bacteria. Biochem. Mol. Biol. Int. 1996, 39, 671–678. [Google Scholar]
- Kitagawa, W.; Tamura, T. A quinolin antibiotic from Phodococcus erthropolis JCM 6824. J. Antibiot. 2008, 61, 680–682. [Google Scholar] [CrossRef]
- Kitagawa, W.; Tamura, T. Three types of antibiotics produced from Phodococcus erthropolis strains. Microbes Environ. 2008, 23, 167–171. [Google Scholar] [CrossRef]
- Gonzalez, M.; Carmen, S.; Miguel, A.; Rao, K.; Sundar, Z-P.; Marmen, M.; Cortes, D. Prenylated benzopyran derivatives from two Polyalthia species. Phytochemistry 1996, 43, 1361–1364. [Google Scholar]
- Zafra-Polo, M.C.; González, M.C.; Tormo, J.R.; Estornell, E.; Cortes, D. Polyalthidin: New prenylated benzopyran inhibitor of the mammalian mitochondrial respiratory chain. J. Nat. Prod. Chem. 1996, 59, 913–916. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; Cabedo, N.; González-Mas, M.C.; Ciavatta, M.L.; Avila, C.; Primo, J. Terretonins E and F, inhibitors of the mitochondrial respiratory chain from the marine-derived fungus Aspergillus insuetus. J. Nat. Prod. 2009, 72, 1348–1351. [Google Scholar] [CrossRef]
- Kurosu, M.; Narayanasamy, P.; Biswas, K.; Dhiman, R.; Crick, D.C. Discovery of 1,4-dihydroxy-2-naphthoate prenyltransferase inhibitors: New drug leads for multidrug-resistant Gram-positive pathogens. J. Med. Chem. 2007, 50, 3973–3975. [Google Scholar] [CrossRef]
- Wayne, L.G.; Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996, 64, 2062–2069. [Google Scholar]
- Wayne, L.G.; Sramek, H.A. Antigenic differences between extracts of actively replicating and synchronized resting cells of Mycobacterium tuberculosis. Infect. Immun. 1979, 24, 363–370. [Google Scholar]
- Wayne, L.G.; Sramek, H.A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1994, 13, 908–912. [Google Scholar]
- Kurosu, M.; Crick, D.C. MenA is a promising drug target for developing novel lead molecules to combat Mycobacterium tuberculosis. Med. Chem. 2009, 5, 197–207. [Google Scholar] [CrossRef]
- Dhiman, R.K.; Mahapatra, S.; Slayden, R.A.; Boyne, M.E.; Lenaerts, A.; Hinshaw, J.C.; Angala, S.K.; Chatterjee, D.; Biswas, K.; Narayanasamy, P.; Kurosu, M.; Crick, D.C. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol. Microbiol. 2009, 72, 85–97. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, H.; Tonge, P.J.; Tan, D.S. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg. Med. Chem. Lett. 2008, 18, 5963–5966. [Google Scholar] [CrossRef]
- Zhang, H.; Tonge, P.J. Enzymatic activity in the crotonase superfamily: The mechanism of the reactions catalyzed by 1,4-dihydroxynaphthoyl-CoA synthase (MenB) and 2-ketocyclohexanecarboxyl-CoA hydrolase (BadI). In 234th ACS National Meeting, Boston, MA, USA, 19–23 August 2007.
- Li, X.; Zhang, H.; Tonge, P.J. Inhibition of 1,4-dihydroxynaphthoyl-CoA synthase (MenB), an enzyme drug target bacterial menaquinone biosynthesis pathway. In 236th ACS National Meeting, Philadelphia, PA, USA, 17–21 August 2008.
- Xu, H.; Graham, M.; Karelis, J.; Walker, S.G.; Peter, J.; Tonge, P.J. Mechanistic studies of MenD, 2-succinyl-5-enoylpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid synthase from Staphylococcus aureus. In 237th ACS National Meeting, Salt Lake City, UT, USA, 22–26 March 2009.
- Hossein, D.G. Stereobiochemical aspects of warfarin isomers for inhibition of enzymatic alkylation of menaquinone-0 to menaquinone-4 in chick liver. Int. J. Vitam. Nutr. Res. 1978, 48, 131–135. [Google Scholar]
- Koul, A.; Vranckx, L.; Dendouga, N.; Balemans, W.; van den Wyngaert, I.; Vergauwen, K.; Göhlmann, H.W.H.; Willebrords, R.; Poncelet, A.; Guillemont, J.; Bald, D.; Andries, K. Diarylquinolines are bactericidal for dormant Mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 2008, 283, 25273–25280. [Google Scholar]
- Cole, S.T.; Alzari, P.M. TB-a new target, a new drug. Science 2005, 307, 214–215. [Google Scholar] [CrossRef]
- Honaker, R.W.; Leistikow, R.L.; Bartek, I.L.; Voskuil, M.I. Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect. Immun. 2009, 77, 3258–3263. [Google Scholar] [CrossRef]
- Farrand, S.K.; Taber, H.W. Physiological effects of menaquinone deficiency in Bacillus subtilis. J. Bacteriol. 1973, 115, 1035–1044. [Google Scholar]
- Lemma, E.; Unden, G.; Kröger, A. Menaquinone is an obligatory component of the chain catalyzing succinate respiration in Bacillus subtilis. Arch. Microbiol. 1990, 155, 62–67. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kurosu, M.; Begari, E. Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets? Molecules 2010, 15, 1531-1553. https://doi.org/10.3390/molecules15031531
Kurosu M, Begari E. Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets? Molecules. 2010; 15(3):1531-1553. https://doi.org/10.3390/molecules15031531
Chicago/Turabian StyleKurosu, Michio, and Eeshwaraiah Begari. 2010. "Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets?" Molecules 15, no. 3: 1531-1553. https://doi.org/10.3390/molecules15031531
APA StyleKurosu, M., & Begari, E. (2010). Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets? Molecules, 15(3), 1531-1553. https://doi.org/10.3390/molecules15031531



