Machine Learning Techniques for Single Nucleotide Polymorphism—Disease Classification Models in Schizophrenia
Abstract
:1. Introduction
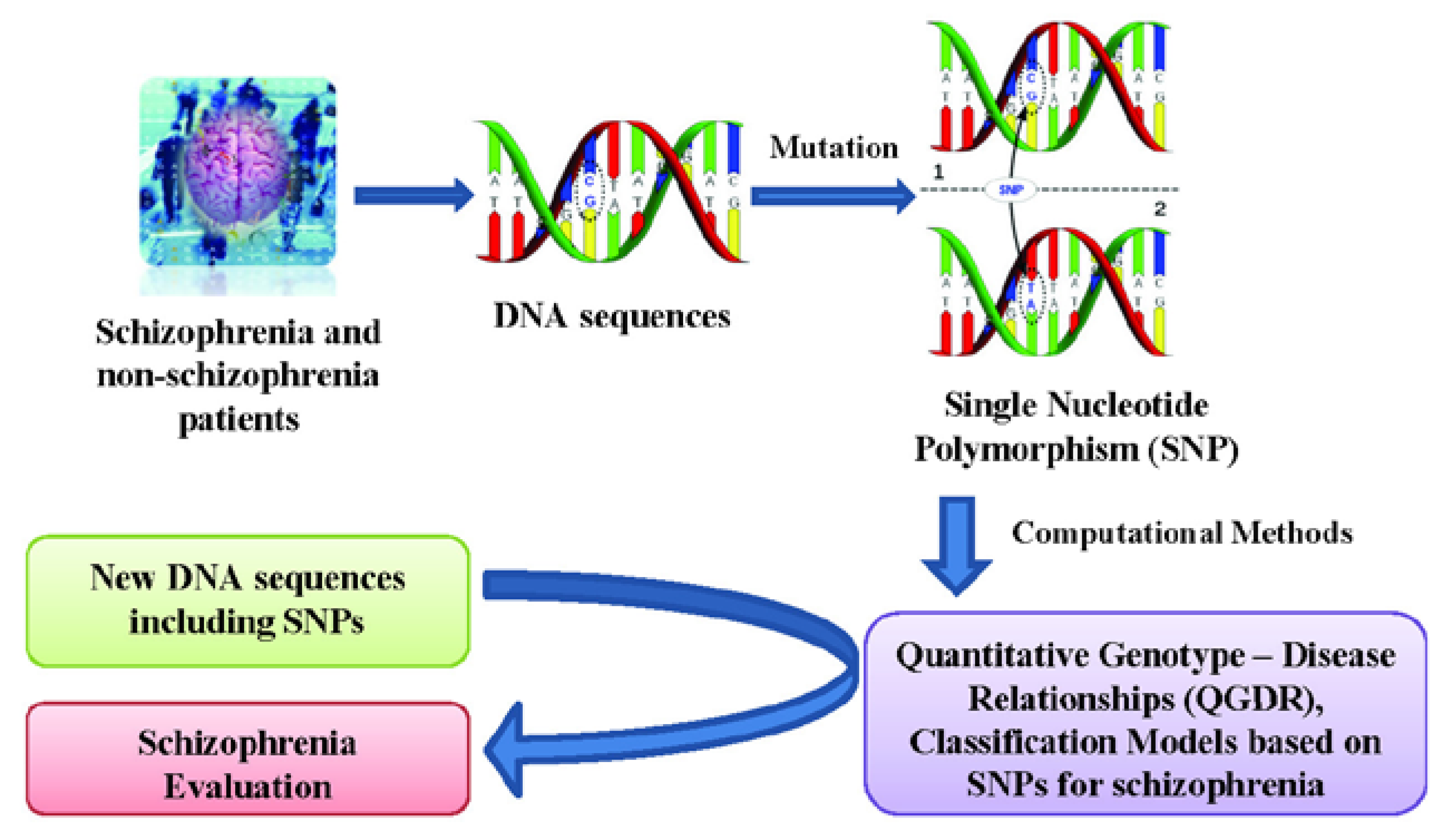
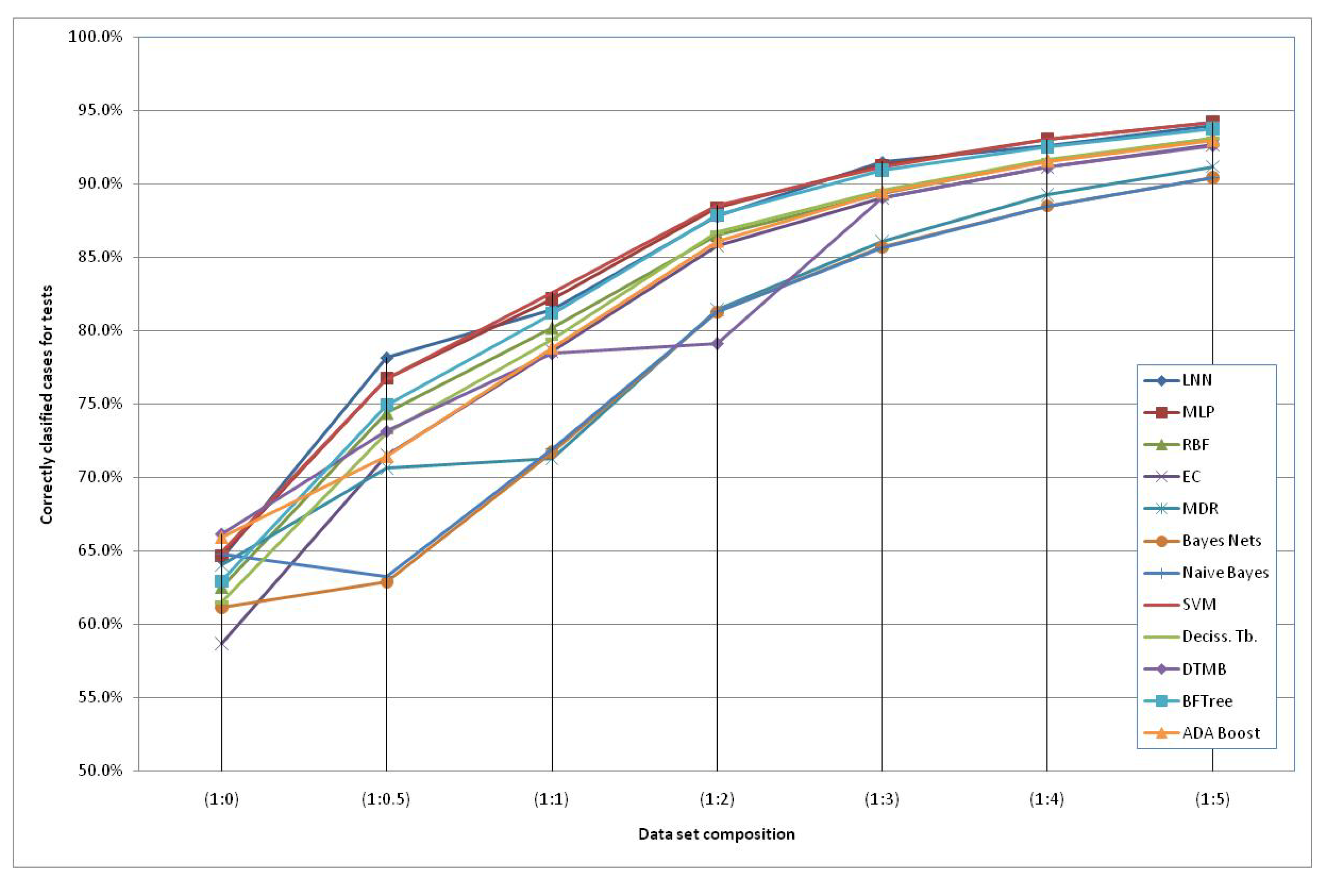
2. Results and Discussion
| Data set | Gene | LNN | MLP | RBF | EC | MDR | Bayes
Nets | Naïve
Bayes | SVM | Decis.
Tb. | DTNB | BFTree | AdaBoost |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (1:0) | DRD3 | 62.9% | 59.5% | 58.9% | 56.6% | 60.0% | 62.5% | 61.6% | 64.8% | 62.2% | 59.5% | 61.3% | 63.4% |
| HTR2A | 62.4% | 62.9% | 63.7% | 57.5% | 64.0% | 61.9% | 66.6% | 65.2% | 61.0% | 62.3% | 62.8% | 63.5% | |
| Both | 64.5% | 64.7% | 62.5% | 58.7% | 64.0% | 61.2% | 64.8% | 64.9% | 61.5% | 66.2% | 62.9% | 65.9% | |
| SNP (1:0.5) | DRD3 | 74.6% | 72.9% | 71.5% | 71.0% | 60.5% | 71.3% | 71.0% | 75.4% | 73.5% | 70.4% | 73.7% | 71.3% |
| HTR2A | 75.9% | 75.5% | 73.6% | 71.7% | 74.2% | 62.2% | 62.9% | 77.4% | 73.2% | 70.9% | 74.5% | 71.4% | |
| Both | 78.2% | 76.8% | 74.4% | 71.5% | 70.7% | 62.9% | 63.3% | 76.8% | 73.1% | 73.2% | 75.0% | 71.4% | |
| SNP (1:1) | DRD3 | 80.5% | 79.5% | 78.5% | 78.2% | 69.8% | 77.9% | 76.2% | 81.4% | 79.6% | 77.1% | 79.4% | 78.6% |
| HTR2A | 80.7% | 81.7% | 80.2% | 78.5% | 71.0% | 71.9% | 72.3% | 83.0% | 79.8% | 76.8% | 81.2% | 78.8% | |
| Both | 81.4% | 82.2% | 80.2% | 78.6% | 71.3% | 71.7% | 72.0% | 82.6% | 79.4% | 78.5% | 81.2% | 78.8% | |
| SNP (1:2) | DRD3 | 87.0% | 86.1% | 85.8% | 85.4% | 79.4% | 84.8% | 83.2% | 87.7% | 86.6% | 80.4% | 86.1% | 85.2% |
| HTR2A | 88.0% | 88.1% | 86.3% | 85.9% | 81.4% | 81.3% | 81.6% | 88.8% | 86.5% | 76.2% | 87.6% | 86.1% | |
| Both | 87.8% | 88.4% | 86.5% | 85.8% | 81.4% | 81.3% | 81.3% | 88.5% | 86.7% | 79.2% | 87.9% | 86.1% | |
| SNP (1:3) | DRD3 | 89.9% | 89.5% | 88.9% | 88.4% | 84.8% | 89.4% | 86.9% | 90.6% | 89.5% | 87.6% | 89.5% | 88.7% |
| HTR2A | 90.4% | 90.7% | 89.3% | 89.1% | 85.9% | 85.7% | 85.9% | 91.4% | 89.7% | 86.5% | 90.3% | 89.4% | |
| Both | 91.5% | 91.3% | 89.3% | 89.1% | 86.1% | 85.7% | 85.6% | 91.2% | 89.5% | 89.1% | 90.9% | 89.4% | |
| SNP (1:4) | DRD3 | 91.9% | 91.7% | 91.3% | 90.9% | 87.4% | 91.5% | 89.2% | 92.5% | 91.6% | 90.3% | 91.5% | 90.7% |
| HTR2A | 92.6% | 92.7% | 91.8% | 91.2% | 88.5% | 88.6% | 88.6% | 93.2% | 91.7% | 88.5% | 92.4% | 91.5% | |
| Both | 92.6% | 93.0% | 91.6% | 91.2% | 89.3% | 88.5% | 88.5% | 93.0% | 91.6% | 91.1% | 92.5% | 91.5% | |
| SNP (1:5) | DRD3 | 93.9% | 93.1% | 93.0% | 92.1% | 88.4% | 92.9% | 90.8% | 93.6% | 93.1% | 91.8% | 92.9% | 92.2% |
| HTR2A | 93.2% | 93.9% | 92.9% | 92.6% | 91.2% | 90.5% | 90.5% | 94.3% | 93.1% | 90.0% | 93.5% | 92.9% | |
| Both | 93.9% | 94.2% | 93.1% | 92.6% | 91.2% | 90.4% | 90.4% | 94.2% | 93.1% | 92.6% | 93.8% | 92.9% |


3. Experimental and Theoretical Section
3.1. Subjects and Genotyping
3.2. Datasets
3.3. QGDR models

4. Conclusions
Acknowledgements
References
- Chinchilla Moreno, A. Las Esquizofrenias. Sus Hechos Y Valores Clínicos Y Terapéuticos; Elsevier Masson: Barcelona, Spain, 2007. [Google Scholar]
- Sham, P. Genetic epidemiology. Br. Med. Bull. 1996, 52, 408–433. [Google Scholar] [CrossRef]
- Sáiz, J.; Fañanás, L. Introducción: Genética y Psiquiatría. Monogr. Psiquiatr. 1998, 10, 1–3. [Google Scholar]
- Meltzer, H.Y.; Matsubara, S.; Lee, J.C. Classification of typical and atypical antipsychotic drugs on the basis of dopamine D-1, D-2 and serotonin2 pKi values. J. Pharmacol. Exp. Ther. 1989, 251, 238–246. [Google Scholar]
- Sokoloff, P.; Levesque, D.; Martres, M.P.; Lannfelt, L.; Diaz, G.; Pilon, C.; Schwartz, J.C. The dopamine D3 receptor as a key target for antipsychotics. Clin. Neuropharmacol. 1992, 15, 456–457. [Google Scholar] [CrossRef]
- Utsunomiya, K.; Shinkai, T.; De Luca, V.; Hwang, R.; Sakata, S.; Fukunaka, Y.; Chen, H.I.; Ohmori, O.; Nakamura, J. Genetic association between the dopamine D3 gene polymorphism (Ser9Gly) and schizophrenia in Japanese populations: evidence from a case-control study and meta-analysis. Neurosci. Lett. 2008, 444, 161–165. [Google Scholar] [CrossRef]
- Suzuki, M.; Hurd, Y.L.; Sokoloff, P.; Schwartz, J.C.; Sedvall, G. D3 dopamine receptor mRNA is widely expressed in the human brain. Brain Res. 1998, 779, 58–74. [Google Scholar] [CrossRef]
- Talkowski, M.E.; Mansour, H.; Chowdari, K.V.; Wood, J.; Butler, A.; Varma, P.G.; Prasad, S.; Semwal, P.; Bhatia, T.; Deshpande, S.; Devlin, B.; Thelma, B.K.; Nimgaonkar, V.L. Novel, replicated associations between dopamine D3 receptor gene polymorphisms and schizophrenia in two independent samples. Biol. Psychiat. 2006, 60, 570–577. [Google Scholar] [CrossRef]
- Dominguez, E.; Loza, M.I.; Padin, F.; Gesteira, A.; Paz, E.; Paramo, M.; Brenlla, J.; Pumar, E.; Iglesias, F.; Cibeira, A.; Castro, M.; Caruncho, H.; Carracedo, A.; Costas, J. Extensive linkage disequilibrium mapping at HTR2A and DRD3 for schizophrenia susceptibility genes in the Galician population. Schizophr. Res. 2007, 90, 123–129. [Google Scholar] [CrossRef]
- den Dunnen, J.T.; Antonarakis, S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000, 15, 7–12. [Google Scholar] [CrossRef]
- Katanforoush, A.; Sadeghi, M.; Pezeshk, H.; Elahi, E. Global haplotype partitioning for maximal associated SNP pairs. BMC Bioinformatics 2009, 10, 269. [Google Scholar] [CrossRef]
- Zhou, N.; Wang, L. Effective selection of informative SNPs and classification on the HapMap genotype data. BMC Bioinformatics 2007, 8, 484. [Google Scholar] [CrossRef]
- Kingman, J. F. Origins of the coalescent. 1974-1982. Genetics 2000, 156, 1461–1463. [Google Scholar]
- Liang, L.; Zollner, S.; Abecasis, G.R. GENOME: a rapid coalescent-based whole genome simulator. Bioinformatics 2007, 23, 1565–1567. [Google Scholar] [CrossRef]
- Wright, F.A.; Huang, H.; Guan, X.; Gamiel, K.; Jeffries, C.; Barry, W.T.; de Villena, F.P.; Sullivan, P.F.; Wilhelmsen, K.C.; Zou, F. Simulating association studies: a data-based resampling method for candidate regions or whole genome scans. Bioinformatics 2007, 23, 2581–2588. [Google Scholar] [CrossRef]
- Balloux, F. EASYPOP (version 1.7): a computer program for population genetics simulations. J. Hered. 2001, 92, 301–302. [Google Scholar] [CrossRef]
- Hey, J. FPG: A Computer Program for Forward Population Genetic Simulation, 2004. Available online: http://lifesci.rutgers.edu/~heylab/HeylabSoftware.htm#FPG/ (accessed on 5 May 2010).
- Hoggart, C.J.; Chadeau-Hyam, M.; Clark, T.G.; Lampariello, R.; Whittaker, J.C.; De Iorio, M.; Balding, D.J. Sequence-level population simulations over large genomic regions. Genetics 2007, 177, 1725–1731. [Google Scholar] [CrossRef]
- Peng, B.; Kimmel, M. simuPOP: a forward-time population genetics simulation environment. Bioinformatics 2005, 21, 3686–3687. [Google Scholar] [CrossRef]
- Edwards, T.L.; Bush, W.S.; Turner, S.D.; Dudek, S.M.; Torstenson, E.S.; Schmidt, M.; Martin, E.; Ritchie, M.D. Generating Linkage Disequilibrium Patterns in Data Simulations using genomeSIMLA. Lect. Notes Comput. Sci. 4973, 24–35. [Google Scholar]
- Li, J.; Chen, Y. Generating samples for association studies based on HapMap data. BMC Bioinformatics 2008, 9, 44. [Google Scholar] [CrossRef]
- Ban, H.J.; Heo, J.Y.; Oh, K.S.; Park, K.J. Identification of Type 2 Diabetes-associated combination of SNPs using Support Vector Machine. BMC Genet. 2010, 11, 26. [Google Scholar]
- Saangyong, U; Dong-Hoi, K.; Young-Woong, K.; Sungwon, C; Jaeyoun, C.; Jin, K. A study on application of single nucleotide polymorphism and machine learning techniques to diagnosis of chronic hepatitis. Expert Systems 2009, 26, 60–69. [Google Scholar] [CrossRef]
- Briggs, F.B.; Ramsay, P.P.; Madden, E.; Norris, J.M.; Holers, V.M.; Mikuls, T.R.; Sokka, T.; Seldin, M.F.; Gregersen, P.K.; Criswell, L.A.; Barcellos, L.F. Supervised machine learning and logistic regression identifies novel epistatic risk factors with PTPN22 for rheumatoid arthritis. Genes Immun. 2010, 11, 199–208. [Google Scholar] [CrossRef]
- Nicodemus, K.K.; Callicott, J.H.; Higier, R.G.; Luna, A.; Nixon, D.C.; Lipska, B.K.; Vakkalanka, R.; Giegling, I.; Rujescu, D.; Clair, D.S.; Muglia, P.; Shugart, Y.Y.; Weinberger, D.R. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum. Genet. 2010, 127, 441–452. [Google Scholar] [CrossRef]
- Devillers, J.; Balaban, A.T. Topological Indices and Related Descriptors in QSAR and QSPR; Gordon and Breach: Amsterdam, The Netherlands, 1999. [Google Scholar]
- Barabasi, A.L.; Bonabeau, E. Scale-free networks. Sci. Am. 2003, 288, 60–69. [Google Scholar] [CrossRef]
- Balaban, A. T.; Basak, S.C.; Beteringhe, A.; Mills, D.; Supuran, C.T. QSAR study using topological indices for inhibition of carbonic anhydrase II by sulfanilamides and Schiff bases. Mol. Divers. 2004, 8, 401–412. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar]
- Barabasi, A.L. Sociology. Network theory-the emergence of the creative enterprise. Science 2005, 308, 639–641. [Google Scholar] [CrossRef]
- González-Díaz, H.; Vilar, S.; Santana, L.; Uriarte, E. Medicinal Chemistry and Bioinformatics – Current Trends in Drugs Discovery with Networks Topological Indices. Curr. Top. Med. Chem. 2007, 7, 1025–1039. [Google Scholar]
- Ferino, G.; Gonzalez-Diaz, H.; Delogu, G.; Podda, G.; Uriarte, E. Using spectral moments of spiral networks based on PSA/mass spectra outcomes to derive quantitative proteome-disease relationships (QPDRs) and predicting prostate cancer. Biochem. Biophys. Res. Commun. 2008, 372, 320–325. [Google Scholar] [CrossRef]
- Gonzalez-Diaz, H.; Gonzalez-Diaz, Y.; Santana, L.; Ubeira, F.M.; Uriarte, E. Proteomics, networks and connectivity indices. Proteomics 2008, 8, 750–778. [Google Scholar] [CrossRef]
- Munteanu, C.R.; Magalhaes, A.L.; Uriarte, E.; Gonzalez-Diaz, H. Multi-target QPDR classification model for human breast and colon cancer-related proteins using star graph topological indices. J. Theor. Biol. 2009, 257, 303–311. [Google Scholar] [CrossRef]
- Vilar, S.; Gonzalez-Diaz, H.; Santana, L.; Uriarte, E. A network-QSAR model for prediction of genetic-component biomarkers in human colorectal cancer. J. Theor. Biol. 2009, 261, 449–458. [Google Scholar] [CrossRef]
- Vilar, S.; Gonzalez-Diaz, H.; Santana, L.; Uriarte, E. QSAR model for alignment-free prediction of human breast cancer biomarkers based on electrostatic potentials of protein pseudofolding HP-lattice networks. J. Comput. Chem. 2008, 29, 2613–2622. [Google Scholar] [CrossRef]
- González-Díaz, H.; Ferino, G.; Prado-Prado, F.J.; Vilar, S.; Uriarte Villares, E.; Pazos, A.; Munteanu, C.R. Protein Graphs in Cancer Prediction. In An Omics Perspective on Cancer Research; Cho, W.C.S., Ed.; Springer Netherlands: Amsterdam, The Netherlands, 2010; doi:10.1007/978-90-481-2675-0_7. [Google Scholar]
- González-Díaz, H.; Ferino, G.; Podda, G.; Uriarte, E. Discriminating Prostate Cancer Patients from control group with connectivity indices. ECSOC 2008, 12, G1:1–G1:10. [Google Scholar]
- Ferino, G.; Delogu, G.; Podda, G.; Uriarte, E.; González-Díaz, H. Quantitative Proteome-Disease Relationships (QPDRs) in Clinical Chemistry: Prediction of Prostate Cancer with Spectral Moments of PSA/MS Star Networks. In Clinical Chemistry Research; Mitchem, B.H., Sharnham, C.L., Eds.; Nova Science Publishers: New York, NY, USA, 2009. [Google Scholar]
- Diederich, J. Artificial Neural Networks: Concept Learning; IEEE Press: Piscataway, NJ, USA, 1990; p. 141. [Google Scholar]
- Byvatov, E.; Schneider, G. Support vector machine applications in bioinformatics. Appl. Bioinformatics 2003, 2, 67–77. [Google Scholar]
- Eberbach, E. Toward a theory of evolutionary computation. Biosystems 2005, 82, 1–19. [Google Scholar] [CrossRef]
- Rowland, J.J. Model selection methodology in supervised learning with evolutionary computation. Biosystems 2003, 72, 187–196. [Google Scholar] [CrossRef]
- Tan, P.-N.; Steinbach, M.; Kumar, V. Introduction to Data Mining; Pearson Addition Wesley: Boston, ML, USA, 2006. [Google Scholar]
- Vapnik, V. Statistical Learning Theory; John Weily and Sons: New York, NY, USA, 1998. [Google Scholar]
- Aguiar Pulido, V.; Seoane Fernández, J.A.; Freire, A.; Munteanu, C.R. Data Mining in Complex Diseases Using Evolutionary Computation. Lect. Notes Comput. Sci. 2009, 5517, 917–924. [Google Scholar] [CrossRef]
- Costas, J.; Torres, M.; Cristobo, I.; Phillips, C.; Carracedo, A. Relative efficiency of the linkage disequilibrium mapping approach in detecting candidate genes for schizophrenia in different European populations. Genomics 2005, 86, 280–286. [Google Scholar] [CrossRef]
- Waikato, T.U.O. Weka Machine Learning Project. Available online: http://www.cs.waikato.ac.nz/ml/weka/ (accessed on 5 May 2010).
- Rosenblatt, F. Principles of Neurodynamics; Perceptrons and The Theory of Brain Mechanisms; Spartan Books: Washington, DC, USA, 1962. [Google Scholar]
- Bishop, C. Neural Networks for Pattern Recognition; Oxford University Press: New York, NY, USA, 1995. [Google Scholar]
- Buhmann, M.D. Radial Basis Functions: Theory and Implementations; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- John, G.H.; Langley, P. Estimating Continuous Distributions in Bayesian Classifiers. In Proceedings of11th Conference on Uncertainty in Artificial Intelligence, Montreal, Quebec, August 18-20 1995; Morgan Kaufmann: San Mateo, CA, USA, 1995; pp. 338–345. [Google Scholar]
- Bouckaert, R.R. Bayesian Networks in Weka; University of Waikato: Tauranga, New Zealand, 2004; Technical report, Computer Science Department. [Google Scholar]
- Kohavi, R. The Power of Decision Tables. In Proceedings of 8th European Conference on Machine Learning, Heraclion, Greece, April 25-27 1995; Springer-Verlag Publisher: London, UK, 1995; pp. 174–189. [Google Scholar]
- Mark Hall, E.F. Combining Naive Bayes and Decision Tables. In Proceedings of the 21st Florida Artificial Intelligence Society Conference (FLAIRS), Coconut Grove, Florida, May 15–17 2008; AAAI Press: Menlo Park, CA, USA, 2008. [Google Scholar]
- Shi, H. Best-first Decision Tree Learning. University of Waikato, Hamilton, New Zealand, 2007. [Google Scholar]
- Freund, Y.; Schapire, R.E. Experiments with a new boosting algorithm. In Proceedings of the Thirteenth International Conference on Machine Learning, Desenzano sul Garda, Italy, June 28 to July 1, 1996; Saitta, L., Ed.; Morgan Kaufmann: San Francisco, CA, 1996; pp. 148–156. [Google Scholar]
- Moore, J.H.; Gilbert, J.C.; Tsai, C.T.; Chiang, F.T.; Holden, T.; Barney, N.; White, B.C. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J. Theor. Biol. 2006, 241, 252–261. [Google Scholar] [CrossRef]
- Cordell, H.J. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 2009, 10, 392–404. [Google Scholar] [CrossRef]
- Greene, C.S.; Sinnott-Armstrong, N.A.; Himmelstein, D.S.; Park, P.J.; Moore, J.H.; Harris, B.T. Multifactor dimensionality reduction for graphics processing units enables genome-wide testing of epistasis in sporadic ALS. Bioinformatics 2010, 26, 694–695. [Google Scholar]
- Cattaert, T.; Urrea, V.; Naj, A.C.; De Lobel, L.; De Wit, V.; Fu, M.; Mahachie John, J.M.; Shen, H.; Calle, M.L.; Ritchie, M.D.; Edwards, T.L.; Van Steen, K. FAM-MDR: a flexible family-based multifactor dimensionality reduction technique to detect epistasis using related individuals. PLoS One 2010, 5, e10304. [Google Scholar]
- He, H.; Oetting, W.S.; Brott, M.J.; Basu, S. Pair-wise multifactor dimensionality reduction method to detect gene-gene interactions in a case-control study. Hum. Hered. 2010, 69, 60–70. [Google Scholar] [CrossRef]
- Kang, S.G.; Lee, H.J.; Choi, J.E.; Park, Y.M.; Park, J.H.; Han, C.; Kim, Y.K.; Kim, S.H.; Lee, M.S.; Joe, S.H.; Jung, I.K.; Kim, L. Association Study between Antipsychotics - Induced Restless Legs Syndrome and Polymorphisms of Dopamine D1, D2, D3, and D4 Receptor Genes in Schizophrenia. Neuropsychobiology 2008, 57, 49–54. [Google Scholar] [CrossRef]
- Vilella, E.; Costas, J.; Sanjuan, J.; Guitart, M.; De Diego, Y.; Carracedo, A.; Martorell, L.; Valero, J.; Labad, A.; De Frutos, R.; Najera, C.; Molto, M.D.; Toirac, I.; Guillamat, R.; Brunet, A.; Valles, V.; Perez, L.; Leon, M.; de Fonseca, F. R.; Phillips, C.; Torres, M. Association of schizophrenia with DTNBP1 but not with DAO, DAOA, NRG1 and RGS4 nor their genetic interaction. J. Psychiatr. Res. 2008, 42, 278–288. [Google Scholar] [CrossRef]
- Yasuno, K.; Ando, S.; Misumi, S.; Makino, S.; Kulski, J.K.; Muratake, T.; Kaneko, N.; Amagane, H.; Someya, T.; Inoko, H.; Suga, H.; Kanemoto, K.; Tamiya, G. Synergistic association of mitochondrial uncoupling protein (UCP) genes with schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007, 144B, 250–253. [Google Scholar] [CrossRef]
- Holland, J.H. Adaptation in Natural and Artificial Systems; University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Darwin, C. On the Origin of Species by Means of Natural Selection; John Murray: London, UK, 1859. [Google Scholar]
- Venturini, G. SIA: A supervised inductive algorithm with genetic search for learning attributes based concepts. In Proceedings of the 6th European Conference on Machine Learning, Vienna, Austria, April 5-7, 1993; Brazdil, P., Ed.; Springer Verlag: Vienna, Austria, 1993; pp. 280–296. [Google Scholar]
- González, A.; Herrera, F. Multi-stage genetic fuzzy systems based on the iterative rule learning approach. Mathware Soft Comput. 1997, 4, 233–249. [Google Scholar]
- McLachlan, G.J.; Do, K.-A.; Ambroise, C. Analyzing Microarray Gene Expression Data . Wiley-Interscience: Hoboken, NJ, USA, 2004. [Google Scholar]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence, Montreal, Canada, August 20-25, 1995; Morgan Kaufmann Publisher: San Francisco, CA, USA, 1995; 2, pp. 1137–1143. [Google Scholar]
- Picard, R.; Cook, D. Cross-Validation of Regression Models. J. Amer. Statist. Assn. 1984, 79, 575–583. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Aguiar-Pulido, V.; Seoane, J.A.; Rabuñal, J.R.; Dorado, J.; Pazos, A.; Munteanu, C.R. Machine Learning Techniques for Single Nucleotide Polymorphism—Disease Classification Models in Schizophrenia. Molecules 2010, 15, 4875-4889. https://doi.org/10.3390/molecules15074875
Aguiar-Pulido V, Seoane JA, Rabuñal JR, Dorado J, Pazos A, Munteanu CR. Machine Learning Techniques for Single Nucleotide Polymorphism—Disease Classification Models in Schizophrenia. Molecules. 2010; 15(7):4875-4889. https://doi.org/10.3390/molecules15074875
Chicago/Turabian StyleAguiar-Pulido, Vanessa, José A. Seoane, Juan R. Rabuñal, Julián Dorado, Alejandro Pazos, and Cristian R. Munteanu. 2010. "Machine Learning Techniques for Single Nucleotide Polymorphism—Disease Classification Models in Schizophrenia" Molecules 15, no. 7: 4875-4889. https://doi.org/10.3390/molecules15074875
APA StyleAguiar-Pulido, V., Seoane, J. A., Rabuñal, J. R., Dorado, J., Pazos, A., & Munteanu, C. R. (2010). Machine Learning Techniques for Single Nucleotide Polymorphism—Disease Classification Models in Schizophrenia. Molecules, 15(7), 4875-4889. https://doi.org/10.3390/molecules15074875






