Regulation of Plant Immunity through Modulation of Phytoalexin Synthesis
Abstract
:1. Introduction
2. Results and Discussion
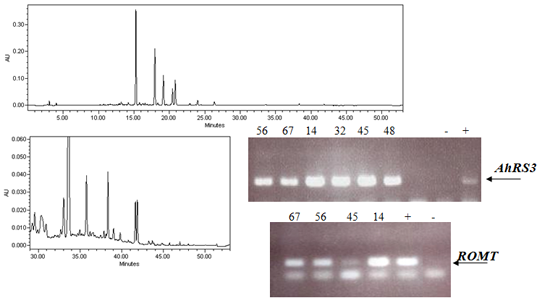
2.1. Expression of Non-Native Stilbenoid Phytoalexins in Soybean Hairy Roots








| Construct | Line | Percent Root Necrosis1 | |
|---|---|---|---|
| None | Control | 83.7 | a |
| CsVMV/AhRS3 | 32 | 42.8 | b |
| CsVMV/G4DT | 98 | 40.5 | bc |
| CsVMV/G4DT | 83 | 39.0 | bc |
| CsVMV/AhRS3 | 48 | 18.6 | cd |
| CsVMV/AhRS3& CsVMV/ROMT | 22 | 6.7 | de |
| CsVMV/AhRS3& CsVMV/ROMT | 45 | 6.5 | de |
| CsVMV/AhRS3& CsVMV/ROMT | 14 | 2.8 | e |
| Actin/AhRS3& CsVMV/ROMT | 67 | 1.8 | de |
| Actin/AhRS3& CsVMV/ROMT | 56 | 0.0 | e |

2.2. Genetic Modulation of the Native Phytoalexin Glyceollin Synthesis in Soybean Hairy Roots

3. Experimental
3.1. Genetic Modification of Soybean Hairy Roots
3.2. Molecular Analysis
3.3. Analysis of Phenylpropanoids in Soybean Tissue
3.4. Evaluation of Fungal Colonization of Hairy Root Cultures
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Islam, A. Fungus resistant transgenic plants: Strategies, progress and lessons learnt. Plant Tissue Cult. Biotech. 2006, 16, 117–138. [Google Scholar]
- Paxton, J.D.; Groth, J. Constraints on pathogens attacking plants. Crit. Rev. Plant Sci. 1994, 13, 77–95. [Google Scholar] [CrossRef]
- VanEtten, H.D.; Mansfield, J.W.; Bailey, J.A.; Farmer, E.E. Two classes of plant antibiotics: Phytoalexins versus phytoanticipins. Plant Cell 1994, 6, 1191–1192. [Google Scholar] [CrossRef]
- VanEtten, H.; Temporini, E.; Wasmann, C. Phytoalexin (and phytoanticipin) tolerance as a virulence trait: Why is it not required by all pathogens? Physiol. Mol. Plant Pathol. 2001, 59, 83–93. [Google Scholar] [CrossRef]
- Kliebenstein, D.J. Secondary metabolites and plant/environment interactions: A view through Arabidopsis thaliana tinged glasses. Plant Cell Environ. 2004, 27, 675–684. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Zheng, Q.A.; Sarma-Mamillapalle, V.K. The phytoalexins from Brassicaceae: Structure, biological activity, synthesis and biosynthesis. Nat. Prod. Commun. 2007, 2, 319–330. [Google Scholar]
- Bednarek, P.; Osbourn, A. Plant-microbe interactions: Chemical diversity in plant defense. Science 2009, 324, 746–748. [Google Scholar] [CrossRef]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef]
- Grobkinsky, D.K.; van der Graaff, E.; Roitsch, T. Phytoalexin transgenics in crop protection—Fairy tale with a happy end? Plant Sci. 2012, 195, 54–70. [Google Scholar] [CrossRef]
- Hain, R.; Reif, H.J.; Krause, E.; Langebartels, R.; Kindl, H.; Vornam, B.; Wiese, W.; Schmelzer, E.; Schreier, P.H.; Stocker, R.H.; et al. Disease resistance results from foreign phytoalexin expression in a novel plant. Nature 1993, 361, 153–156. [Google Scholar] [CrossRef]
- Papadopoulou, K.; Melton, R.E.; Leggett, M.; Daniels, M.J.; Osbourn, A.E. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 1999, 96, 12923–12928. [Google Scholar] [CrossRef]
- Essenberg, M. Prospects for strengthening plant defenses through phytoalexin engineering. Physiol. Mol. Plant Pathol. 2001, 59, 71–81. [Google Scholar] [CrossRef]
- Campbell, M.A.; Fitzgerald, H.A.; Ronald, P.C. Engineering pathogen resistance in crop plants. Transgenic Res. 2002, 11, 599–613. [Google Scholar] [CrossRef]
- Jeandet, P.; Douillet, A.C.; Debord, S.; Sbaghi, M.; Bessis, R.; Adrian, M. Phytoalexins from the Vitaceae: Biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J. Agric. Food Chem. 2002, 50, 2731–2741. [Google Scholar]
- Jeandet, P.; Delaunois, B.; Conreux, A.; Donnez, D.; Nuzzo, V.; Cordelier, S.; Clement, C.; Courot, E. Biosynthesis, metabolism, molecular engineering and biological functions of stilbene phytoalexins in plants. Biofactors 2010, 36, 331–341. [Google Scholar] [CrossRef]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Widholm, J.M. Genetic engineering of plant; disease resistance by modification of the phenylpropanoid pathway. Plant Biosyst. 2005, 139, 20–23. [Google Scholar] [CrossRef]
- Aharoni, A.; Galili, G. Metabolic engineering of the plant primary-secondary metabolism interface. Curr. Opin. Biotechnol. 2011, 22, 239–244. [Google Scholar] [CrossRef]
- Jeandet, P.; Clement, C.; Courot, E.; Cordelier, S. Modulation of Phytoalexin Biosynthesis in Engineered Plants for Disease Resistance. Int. J. Mol. Sci. 2013, 14, 14136–14170. [Google Scholar] [CrossRef]
- Lozovaya, V.V.; Lygin, A.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Biochemical response of soybean roots to Fusarium solani f.sp. glycines infection. Crop Sci. 2004, 44, 819–826. [Google Scholar] [CrossRef]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Ulanov, A.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 2007, 225, 665–679. [Google Scholar] [CrossRef]
- Lygin, A.V.; Li, S.; Vittal, R.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. The importance of phenolic metabolism to limit the growth of Phakopsora pachyrhizi. Phytopathology 2009, 99, 1412–1420. [Google Scholar] [CrossRef]
- Lygin, A.V.; Hill, C.B.; Zernova, O.V.; Crull, L.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. Response of Soybean Pathogens to Glyceollin. Phytopathology 2010, 100, 897–903. [Google Scholar]
- Lygin, A.V.; Zernova, O.V.; Hill, C.B.; Kholina, N.A.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. Glyceollin is an important component of soybean plant defense against Phytophthora sojae and Macrophomina phaseolina. Phytopathology 2013, 103, 984–994. [Google Scholar] [CrossRef]
- Pervaiz, S.; Holme, A.L. Resveratrol: Its biologic targets and functional activity. Antioxid. Redox Signal. 2009, 11, 2851–2897. [Google Scholar] [CrossRef]
- Fulda, S. Resveratrol and derivatives for the prevention and treatment of cancer. Drug Discov. Today 2010, 15, 757–765. [Google Scholar] [CrossRef]
- Hain, R.; Bieseer, B.; Kindl, H.; Schroeder, G.; Stoecker, R. Expresshion of a stilbene synthase gene in Nicotiana-tabacum results in synthesis of the phytoalexin resveratrol. Plant Mol. Biol. 1990, 15, 325–336. [Google Scholar] [CrossRef]
- Thomzik, J.E.; Stenzel, K.; Stocker, R.; Schreier, P.H.; Hain, R.; Stahl, D.J. Synthesis of a grapevine phytoalexin in transgenic tomatoes (Lycopersicon esculentum Mill.) conditions resistance against Phytophthora infestans. Physiol. Mol. Plant Pathol. 1997, 51, 265–278. [Google Scholar] [CrossRef]
- Leckband, G.; Lorz, H. Transformation and expression of a stilbene synthase gene of Vitis Vinifera L. in barley and wheat for increased fungal resistance. Theor. Appl. Genet. 1998, 96, 1004–1012. [Google Scholar] [CrossRef]
- Hipskind, J.D.; Paiva, N.L. Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol. Plant Microbe Interact. 2000, 13, 551–562. [Google Scholar] [CrossRef]
- Coutos-Thevenot, P.; Poinssot, B.; Bonomelli, A.; Yean, H.; Breda, C.; Buffard, D.; Esnault, R.; Hain, R.; Boulay, M. In vitro tolerance to Botrytis cinerea of grapevine 41B rootstock in transgenic plants expressing the stilbene synthase VST1 gene under the control of a pathogen-inducible PR 10 promoter. J. Exp. Botany 2001, 52, 901–910. [Google Scholar] [CrossRef]
- Delaunois, B.; Cordelier, S.; Conreux, A.; Clément, C.; Jeandet, P. Molecular engineering of resveratrol in plants. Plant Biotech. J. 2009, 7, 2–12. [Google Scholar] [CrossRef]
- Pezet, R.; Pont, V. Ultrastractural observation of pterostilbene fungitoxity in dormant conidia of Botrytis-cinerea pers. J. Phytopathol. 1990, 129, 19–30. [Google Scholar] [CrossRef]
- Pezet, R.; Gindro, K.; Viret, O.; Spring, J.-L. Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol. Mol. Plant Pathol. 2004, 65, 297–303. [Google Scholar] [CrossRef]
- Lygin, A.V.; Hill, C.B.; Pawlowski, M.; Zernova, O.V.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. Inhibitory effects of stilbenes on soybean pathogen growth. Phytopathology 2014, in press. [Google Scholar]
- Rimando, A.M.; Pan, Z.; Polashok, J.J.; Dayan, F.E.; Mizuno, C.S.; Snook, M.E.; Liu, C.-J.; Baerson, S.R. In planta production of the highly potent resveratrol analogue pterostilbene via stilbene synthase and 0-methyltransferase co-expression. Plant Biotechnol. J. 2011, 10, 269–283. [Google Scholar]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef]
- Akashi, T.; Sasaki, K.; Aoki, T.; Ayabe, S.; Yazaki, K. Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 2009, 149, 683–693. [Google Scholar]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Isoflavonoid accumulation in soybean hairy roots upon treatment with Fusariun solani. Plant Physiol. Biochem. 2004, 42, 671–679. [Google Scholar] [CrossRef]
- Lim, J.; Song, J.; Chung, I.; Yu, C. Resveratrol synthase transgene expression and accumulation of resveratrol glycoside in Rehmannia glutinosa) ubiqiutin promoter for selection. Mol. Breed. 2005, 16, 219–233. [Google Scholar] [CrossRef]
- Schmidlin, L.; Poutaraud, A.; Claudel, P.; Mestre, P.; Prado, E.; Santos-Rosa, M.; Wiedemann-Merdinoglu, S.; Karst, F.; Merdinoglu, D.; Hugueney, P. A stress-inducible resveratrol o-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 2008, 148, 1630–1639. [Google Scholar] [CrossRef]
- Dellaporta, S.L. Plant DNA miniprep version 2.1–2.3. In The Maize Handbook; Freeling, M., Walbot, V., Eds.; Springer-Verlag: New York, NY, USA, 1993; pp. 522–525. [Google Scholar]
- Zernova, O.V.; Lygin, A.V.; Widholm, J.M.; Lozovaya, V.V. Modification of isoflavones in soybean seeds via expression of multiple phenolic biosynthetic genes. Plant Physiol. Biochem. 2009, 47, 769–777. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2014 by the authors. licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zernova, O.V.; Lygin, A.V.; Pawlowski, M.L.; Hill, C.B.; Hartman, G.L.; Widholm, J.M.; Lozovaya, V.V. Regulation of Plant Immunity through Modulation of Phytoalexin Synthesis. Molecules 2014, 19, 7480-7496. https://doi.org/10.3390/molecules19067480
Zernova OV, Lygin AV, Pawlowski ML, Hill CB, Hartman GL, Widholm JM, Lozovaya VV. Regulation of Plant Immunity through Modulation of Phytoalexin Synthesis. Molecules. 2014; 19(6):7480-7496. https://doi.org/10.3390/molecules19067480
Chicago/Turabian StyleZernova, Olga V., Anatoli V. Lygin, Michelle L. Pawlowski, Curtis B. Hill, Glen L. Hartman, Jack M. Widholm, and Vera V. Lozovaya. 2014. "Regulation of Plant Immunity through Modulation of Phytoalexin Synthesis" Molecules 19, no. 6: 7480-7496. https://doi.org/10.3390/molecules19067480
APA StyleZernova, O. V., Lygin, A. V., Pawlowski, M. L., Hill, C. B., Hartman, G. L., Widholm, J. M., & Lozovaya, V. V. (2014). Regulation of Plant Immunity through Modulation of Phytoalexin Synthesis. Molecules, 19(6), 7480-7496. https://doi.org/10.3390/molecules19067480