Inhibitory Effects of Neochamaejasmin B on P-Glycoprotein in MDCK-hMDR1 Cells and Molecular Docking of NCB Binding in P-Glycoprotein
Abstract
:1. Introduction

2. Results and Discussion
2.1. Cytotoxicity of NCB for in Vitro Incubation
2.2. R-123 Transport Studies
| Compound | Papp (cm/s) × 10−7 | Efflux Ratio (Papp BL-AP/PappAP-BL) | |
|---|---|---|---|
| BL-AP | AP-BL | ||
| R-123 | 23.9 ± 1.54 | 3.63 ± 0.31 | 6.59 ± 0.14 |
| R-123 + 50 μM NCB | 8.67 ± 1.02 | 3.59 ± 0.33 | 2.41 ± 0.062 |

2.3. R-123 Accumulation Assay


2.4. Ki and Ki’ Assay


2.5. Quantitative Determination of ABCB1 mRNA Expression

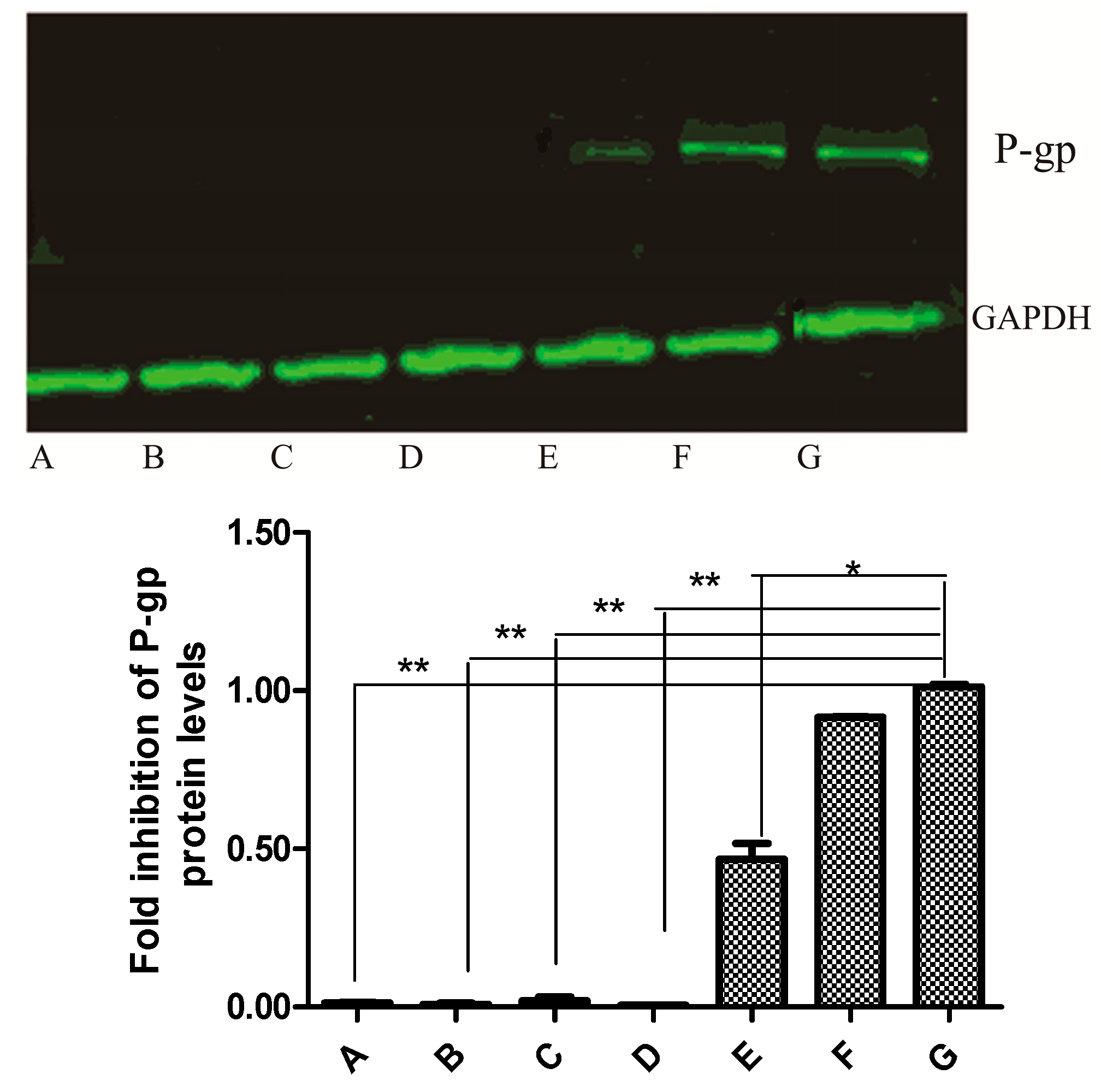
2.6. Western Blot Analysis
2.7. Binding Selectivity Studies Based on Molecular Docking



2.8. Discussion
3. Experimental Section
3.1. Materials
3.2. Cell Culture
3.3. In Vitro Cytotoxicity Assays
3.4. R-123 Transport Experiments
3.5. Transport Experiments Data Analyses
3.6. Cellular Accumulation
3.6.1. R-123 Accumulation Assay
3.6.2. R-123 Accumulation Data Analyses
Ki and Ki’ Assay
3.7. Determination of ABCB1 mRNA Gene Expression
3.8. Analysis of P-gp Protein Expression by Western Blot
3.9. Binding Selectivity Studies Based on Molecular Docking
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Asada, Y.; Sukemori, A.; Watanabe, T.; Malla, K.J.; Yoshikawa, T.; Li, W.; Koike, K.; Chen, C.H.; Akiyama, T.; Qian, K.; et al. Stelleralides A-C, novel potent anti-HIV daphnane-type diterpenoids from Stellera chamaejasme L. Org. Lett. 2011, 13, 2904–2907. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, D. Biflavanones, flavonoids, and coumarins from the roots of Stellera chamaejasme and their antiviral effect on hepatitis B virus. Chem. Biodivers. 2008, 5, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Duan, H.; Wang, Y.; Liu, K.; Jiang, P.; Qu, Z.; Yagasaki, K.; Zhang, G. Inhibitory effects of Lang-du extract on the in vitro and in vivo growth of melanoma cells and its molecular mechanisms of action. Cytotechnology 2010, 62, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.L.; Liu, S.Q.; Cao, H.; Zhao, L.L.; Li, J.; Li, S.Y. Acaricidal activities of extracts of Stellera chamaejasme against Tetranychus viennensis (Acari: Tetranychidae). J. Econ. Entomol. 2004, 97, 1912–1916. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.H.; Qin, G.W.; Li, X.Y.; Xu, R.S. New biflavanones and bioactive compounds from Stellera chamaejasme L. Yao Xue Xue Bao 2001, 36, 669–671. [Google Scholar] [PubMed]
- Tang, X.; Hou, T. Isolation and identification of 2-isopropyl-5-methylphenol from Stellera chamaejasme and its insecticidal activity against Aphis craccivora and Pieris rapae. Nat. Prod. Res. 2011, 25, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.T.; Lin, S.S.; Wang, C.K.; Lee, Y.B.; Chen, K.S.; Fong, Y.; Shih, Y.W. Acacetin inhibits the invasion and migration of human non-small cell lung cancer A549 cells by suppressing the p38alpha MAPK signaling pathway. Mol. Cell Biochem. 2011, 350, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ye, S.; Qin, R.; Deng, Y.; Li, C.P. Effect of Chinese herbal medicine extracts on cell-mediated immunity in a rat model of tuberculosis induced by multiple drug-resistant bacilli. Mol. Med. Rep. 2013, 8, 227–232. [Google Scholar] [PubMed]
- Lou, Y.; Hu, H.; Liu, Y.; Yu, Q.; Li, L.; Ping, L.; Yu, L.; Jiang, H.; Zeng, S. Determination of chamaechromone in rat plasma by liquid chromatography-tandem mass spectrometry: Application to pharmacokinetic study. J. Pharm. Biomed. Anal. 2011, 55, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Zheng, J.; Wang, B.; Zhang, X.; Zhang, X.; Zeng, S. Metabolites characterization of chamaechromone in vivo and in vitro by using ultra-performance liquid chromatography/Xevo G2 quadrupole time-of-flight tandem mass spectrometry. J. Ethnopharmacol. 2014, 151, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Bhaskaran, N.; Babcook, M.A.; Fu, P.; Maclennan, G.T.; Gupta, S. Apigenin inhibits prostate cancer progression in TRAMP mice via targeting PI3K/Akt/FoxO pathway. Carcinogenesis 2014, 35, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Juvale, K.; Stefan, K.; Wiese, M. Synthesis and biological evaluation of flavones and benzoflavones as inhibitors of BCRP/ABCG2. Eur. J. Med. Chem. 2013, 67C, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhao, L.; Wang, H.; Wang, Y.; Pan, D.; Yao, J.; Li, Z.; Wu, G.; Guo, Q. Oroxylin A reverses P-glycoprotein-mediated multidrug resistance of MCF7/ADR cells by G2/M arrest. Toxicol. Lett. 2013, 219, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Schmidt, K.; Nelson, F.R.; Zelesky, V.; Troutman, M.D.; Feng, B. The effect of breast cancer resistance protein and P-glycoprotein on the brain penetration of flavopiridol, imatinib mesylate (Gleevec), prazosin, and2-methoxy-3-(4-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)phenyl)propanoic acid (PF-407288) in mice. Drug Metab. Dispos. 2009, 37, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, H.; Bruggisser, R.; Schaffner, W.; Bogman, K.; Botomino, A.; Drewe, J. Transport of amentoflavone across the blood-brain barrier in vitro. Planta Med. 2002, 68, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, S. Advances in the MDCK-MDR1 cell model and its applications to screen drug permeability. Yao Xue Xue Bao 2008, 43, 559–564. [Google Scholar] [PubMed]
- Al-Shawi, M.K.; Senior, A.E. Characterization of the adenosine triphosphatase activity of Chinese hamster P-glycoprotein. J. Biol. Chem. 1993, 268, 4197–4206. [Google Scholar] [PubMed]
- Aberg, M.; Wickstrom, M.; Siegbahn, A. Simvastatin induces apoptosis in human breast cancer cells in a NFκB-dependent manner and abolishes the anti-apoptotic signaling of TF/FVIIa and TF/FVIIa/FXa. Thromb. Res. 2008, 122, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Tetsuro, I.; Mitsuzi, Y. The antitumor activities of gnidimacrin isolated from Stellera chamaejasme L. Zhonghua Zhong Liu Za Zhi 1995, 17, 24–26. [Google Scholar] [PubMed]
- Zhang, S.; Sagawa, K.; Arnold, R.D.; Tseng, E.; Wang, X.; Morris, M.E. Interactions between the flavonoid biochanin A and P-glycoprotein substrates in rats: In vitro and in vivo. J. Pharm. Sci. 2010, 99, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Oudjeriouat, N.; Moreau, Y.; Santimone, M.; Svensson, B.; Marchis-Mouren, G.; Desseaux, V. On the mechanism of α-amylase. Eur. J. Biochem. 2003, 270, 3871–3879. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Sohier, J.S.; Feller, G. Kinetics and energetics of ligand binding determined by microcalorimetry: insights into active site mobility in a psychrophilic α-amylase. J. Mol. Biol. 2006, 358, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Lovelace, L.L.; Gibson, L.M.; Lebioda, L. Cooperative inhibition of human thymidylate synthase by mixtures of active site binding and allosteric inhibitors. Biochemistry 2007, 46, 2823–2830. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; McClendon, C.L.; Jacobson, M.P.; Wells, J.A. Substrate and inhibitor-induced dimerization and cooperativity in caspase-1 but not caspase-3. J. Biol. Chem. 2013, 288, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F. Cystic fibrosis transmembrane conductance regulator (CFTR). Br. Med. Bull. 1992, 48, 754–765. [Google Scholar] [PubMed]
- Crozat, E.; Grainge, I. FtsK DNA translocase: The fast motor that knows where it’s going. Chembiochem 2010, 11, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Bocian, W.; Borowicz, P.; Mikolajczyk, J.; Sitkowski, J.; Tarnowska, A.; Bednarek, E.; Glabski, T.; Tejchman-Malecka, B.; Bogiel, M.; Kozerski, L. NMR structure of biosynthetic engineered human insulin monomer B31(Lys)-B32(Arg) in water/acetonitrile solution. Comparison with the solution structure of native human insulin monomer. Biopolymers 2008, 89, 820–830. [Google Scholar]
- Lo, A.; Burckart, G.J. P-glycoprotein and drug therapy in organ transplantation. J. Clin. Pharmacol. 1999, 39, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, S.; Nabekura, T.; Takahashi, T.; Nakamura, Y.; Sakamoto, H.; Tano, H.; Hirai, M.; Tsukahara, G. Structure-activity relationships of the inhibitory effects of flavonoids on P-glycoprotein-mediated transport in KB-C2 cells. Biol. Pharm. Bull. 2005, 28, 2274–2278. [Google Scholar] [CrossRef] [PubMed]
- Boumendjel, A.; di Pietro, A.; Dumontet, C.; Barron, D. Recent advances in the discovery of flavonoids and analogs with high-affinity binding to P-glycoprotein responsible for cancer cell multidrug resistance. Med. Res. Rev. 2002, 22, 512–529. [Google Scholar] [CrossRef] [PubMed]
- Sheu, M.T.; Liou, Y.B.; Kao, Y.H.; Lin, Y.K.; Ho, H.O. A quantitative structure-activity relationship for the modulation effects of flavonoids on p-glycoprotein-mediated transport. Chem. Pharm. Bull. 2010, 58, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Naito, M.; Oh-hara, T.; Yamazaki, A.; Danki, T.; Tsuruo, T. Reversal of multidrug resistance by an immunosuppressive agent FK-506. Cancer Chemother. Pharmacol. 1992, 29, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, S.S.; Feng, L.Y.; Zhang, D.Y.; Lin, N.M.; Zhang, L.H.; Pan, J.P.; Wang, J.B.; Li, J. In vitro anti-cancer activity of chamaejasmenin B and neochamaejasmin C isolated from the root of Stellera chamaejasme L. Acta Pharmacol. Sin. 2013, 34, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Yang, Q.; Chen, Y.; Weng, X.; Wang, Y.; Zhu, X. Comparative study on tumor cell apoptosis in vitro induced by extracts of Stellera chamaejasme. Zhongguo Zhong Yao Za Zhi 2012, 37, 1440–1444. [Google Scholar] [PubMed]
- Razzaque, M.S.; Koji, T.; Kumatori, A.; Taguchi, T. Cisplatin-induced apoptosis in human proximal tubular epithelial cells is associated with the activation of the Fas/Fas ligand system. Histochem. Cell Biol. 1999, 111, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Uslu, R.; Borsellino, N.; Frost, P.; Garban, H.; Ng, C.P.; Mizutani, Y.; Belldegrun, A.; Bonavida, B. Chemosensitization of human prostate carcinoma cell lines to anti-fas-mediated cytotoxicity and apoptosis. Clin. Cancer Res. 1997, 3, 963–972. [Google Scholar] [PubMed]
- Kozlowski, M.; Kowalczuk, O.; Sulewska, A.; Dziegielewski, P.; Lapuc, G.; Laudanski, W.; Niklinska, W.; Chyczewski, L.; Niklinski, J.; Laudanski, J. Serum soluble Fas ligand (sFasL) in patients with primary squamous cell carcinoma of the esophagus. Folia Histochem Cytobiol. 2007, 45, 199–204. [Google Scholar] [PubMed]
- Niwa, M.; Tatematsu, H.; Liu, G.Q. Isolation and structures of two new C-3/C-3''-biflavanones, neochamaejasmin A and neochamaejasmin B. Chem. Lett. 1984, 53, 539–542. [Google Scholar] [CrossRef]
- Gu, W.J.; Liu, H.L. Induction of pancreatic cancer cell apoptosis, invasion, migration, and enhancement of chemotherapy sensitivity of gemcitabine, 5-FU, and oxaliplatin by hnRNP A2/B1 siRNA. Anticancer Drugs 2013, 24, 566–576. [Google Scholar] [PubMed]
- Zhang, Y.; Zhou, T.; Duan, J.; Xiao, Z.; Li, G.; Xu, F. Inhibition of P-glycoprotein and Glutathione S-transferase-pi mediated resistance by fluoxetine in MCF-7/ADM cells. Biomed. Pharmacother. 2013, 67, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, F.; Wu, X.; Gu, Y.; Ai, H.; Zheng, Y.; Li, Y.; Zhang, X.; Hao, G.; Sun, J.; et al. 20(S)-ginsenoside Rh2 noncompetitively inhibits P-glycoprotein in vitro and in vivo: A case for herb-drug interactions. Drug Metab. Dispos. 2010, 38, 2179–2187. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miyata, M.; Zaima, C.; Furuishi, T.; Fukami, T.; Kugawa, F.; Tomono, K. Blood-brain barrier transport of naloxone does not involve P-glycoprotein-mediated efflux. J. Pharm. Sci. 2010, 99, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Berger, V.; Gabriel, A.F.; Sergent, T.; Trouet, A.; Larondelle, Y.; Schneider, Y.J. Interaction of ochratoxin A with human intestinal Caco-2 cells: Possible implication of a multidrug resistance-associated protein (MRP2). Toxicol. Lett. 2003, 140–141, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.M.; Lee, S.C.; Shook, S.; Pappone, P.A. ATP and beta-adrenergic stimulation enhance voltage-gated K current inactivation in brown adipocytes. Am. J. Physiol. Cell Physiol. 2000, 279, C1847–C1858. [Google Scholar] [PubMed]
- Intekhab-Alam, N.Y.; White, O.B.; Getting, S.J.; Petsa, A.; Knight, R.A.; Chowdrey, H.S.; Townsend, P.A.; Lawrence, K.M.; Locke, I.C. Urocortin protects chondrocytes from NO-induced apoptosis: A future therapy for osteoarthritis? Cell Death Dis. 2013, 4, e717. [Google Scholar] [CrossRef] [PubMed]
- Molnar, J.; Engi, H.; Hohmann, J.; Molnar, P.; Deli, J.; Wesolowska, O.; Michalak, K.; Wang, Q. Reversal of multidrug resitance by natural substances from plants. Curr. Top. Med. Chem. 2010, 10, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.E.; Cho, J.K.; Curtis-Long, M.J.; Ryu, H.W.; Kim, J.H.; Kim, H.J.; Yuk, H.J.; Kim, D.W.; Park, K.H. Inhibitory evaluation of sulfonamide chalcones on β-secretase and acylcholinesterase. Molecules 2012, 18, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Sivaganensan, M.; Varma, M.; Haugland, R.A. Comparison of Enterococcus quantitative polymerase chain reaction analysis results from fresh and marine waters on two real-time instruments. Anal. Biochem. 2012, 430, 68–74. [Google Scholar] [CrossRef]
- Funk, A.J.; McCullumsmith, R.E.; Haroutunian, V.; Meador-Woodruff, J.H. Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology 2012, 37, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Madeira da Silva, L.; Vandepas, L.; Bianco, S.D. Mutagenesis and analysis of genetic mutations in the GC-rich KISS1 receptor sequence identified in humans with reproductive disorders. J. Vis. Exp. 2011, e2897. [Google Scholar] [CrossRef]
- Aller, S.G.; Yu, J.; Ward, A.; Weng, Y.; Chittaboina, S.; Zhuo, R.; Harrell, P.M.; Trinh, Y.T.; Zhang, Q.; Urbatsch, I.L.; Chang, G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 2009, 323, 1718–1722. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Han, S.; Cao, Y.; Chen, J.Z. The agonist binding mechanism of human CB2 receptor studied by molecular dynamics simulation, free energy calculation and 3D-QSAR studies. Yao Xue Xue Bao 2013, 48, 1436–1449. [Google Scholar] [PubMed]
- Sample Availability: Sample of the compound NCB is available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Hu, H.; Wang, X.; Yu, L.; Jiang, H.; Chen, J.; Lou, Y.; Zeng, S. Inhibitory Effects of Neochamaejasmin B on P-Glycoprotein in MDCK-hMDR1 Cells and Molecular Docking of NCB Binding in P-Glycoprotein. Molecules 2015, 20, 2931-2948. https://doi.org/10.3390/molecules20022931
Pan L, Hu H, Wang X, Yu L, Jiang H, Chen J, Lou Y, Zeng S. Inhibitory Effects of Neochamaejasmin B on P-Glycoprotein in MDCK-hMDR1 Cells and Molecular Docking of NCB Binding in P-Glycoprotein. Molecules. 2015; 20(2):2931-2948. https://doi.org/10.3390/molecules20022931
Chicago/Turabian StylePan, Lanying, Haihong Hu, Xiangjun Wang, Lushan Yu, Huidi Jiang, Jianzhong Chen, Yan Lou, and Su Zeng. 2015. "Inhibitory Effects of Neochamaejasmin B on P-Glycoprotein in MDCK-hMDR1 Cells and Molecular Docking of NCB Binding in P-Glycoprotein" Molecules 20, no. 2: 2931-2948. https://doi.org/10.3390/molecules20022931
APA StylePan, L., Hu, H., Wang, X., Yu, L., Jiang, H., Chen, J., Lou, Y., & Zeng, S. (2015). Inhibitory Effects of Neochamaejasmin B on P-Glycoprotein in MDCK-hMDR1 Cells and Molecular Docking of NCB Binding in P-Glycoprotein. Molecules, 20(2), 2931-2948. https://doi.org/10.3390/molecules20022931








