Synthesis, Biological Evaluation and Molecular Docking of Certain Sulfones as Potential Nonazole Antifungal Agents
Abstract
:1. Introduction

2. Results and Discussion
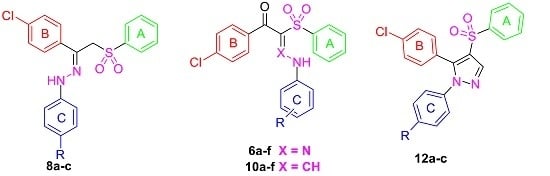
2.1. Chemistry



2.2. Biological Data
2.2.1. In Vitro Antifungal Activities

| Comp. | Invasive Fungi | Dermatophytic Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| CaR | Ca | Ct | Cp | Af | An | Tm | Mc | |
| 6a | 14.2 ± 0.58 | 12.4 ± 0.63 | 11.5 ± 0.58 | 12.4 ± 0.63 | 11.2 ± 0.63 | 11.9 ± 0.63 | 9.3 ± 0.63 | 10.2 ± 0.63 |
| 8a | 18.2 ± 0.72 | 20.3 ± 0.63 | 19.4 ± 0.58 | 20.6 ± 0.63 | NA | NA | NA | NA |
| 10a | NA | 12.3 ± 0.63 | 14.1 ± 0.63 | 15.2 ± 0.58 | 10.6 ± 0.72 | 9.3 ± 0.58 | 10.1 ± 0.63 | 12.1 ± 0.58 |
| 10b | 19.6 ± 0.63 | 20.9 ± 0.72 | 21.2 ± 0.45 | 22.4 ± 0.58 | NA | NA | NA | NA |
| 10e | NA | NA | NA | NA | 20.3 ± 0.63 | 18.2 ± 0.63 | 17.1 ± 0.63 | 18.9 ± 0.63 |
| 12a | NA | NA | NA | NA | 19.3 ± 0.72 | 18.1 ± 0.58 | 18.2 ± 0.72 | 20.1 ± 0.58 |
| Flu | NA | 16.6 ± 0.58 | 15.4 ± 0.58 | 17.2 ± 0.72 | 19.6 ± 0.58 | 16.3 ± 0.63 | 13.6 ± 0.72 | 15.4 ± 0.63 |
| Comp. | Invasive Fungi | Dermatophytic Fungi | PC3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CaR | Ca | Ct | Cp | Af | An | Tm | Mc | IC50 | |
| 6a | 1.57 | 1.57 | 1.57 | 1.57 | 1.57 | 1.57 | 3.14 | 1.57 | 38.86 |
| (24.75) | (24.75) | (24.75) | (24.75) | (24.75) | (24.75) | (12.38) | (24.75) | ||
| 8a | 0.81 | 0.20 | 0.41 | 0.20 | NA | NA | NA | NA | 37.36 |
| (46.12) | (>100) | (91.12) | (>100) | ||||||
| 10a | NA | 1.57 | 1.57 | 1.57 | 1.57 | 3.14 | 1.57 | 1.57 | 35.14 |
| (22.38) | (22.38) | (22.38) | (22.38) | (11.19) | (22.38) | (22.38) | |||
| 10b | 0.19 | 0.19 | 0.19 | 0.19 | NA | NA | NA | NA | 37.09 |
| (>100) | (>100) | (>100) | (>100) | ||||||
| 10e | NA | NA | NA | NA | 0.16 | 0.66 | 0.66 | 0.66 | 38.73 |
| (>100) | (58.68) | (58.68) | (58.68) | ||||||
| 12a | NA | NA | NA | NA | 0.20 | 0.79 | 0.79 | 0.20 | 28.90 |
| (>100) | (36.6) | (36.60) | (>100) | ||||||
| Fluconazole | NA | 1 | 1 | 1 | 2.6 | 2 | 2 | 2 | |
2.2.2. In Vitro Cytotoxicity
2.3. Computational Study
2.3.1. Docking


| Compound | CDOCKER Interaction Energy (kcal/mol) |
|---|---|
| 6a | −42.431 |
| 8a | −36.214 |
| 10a | −34.873 |
| 10b | −39.229 |
| Fluconazole | −40.374 |

2.3.2. ADME and Molecular Property Prediction
| Comp. | AlogP98 a | PSA b | Solubility Level c | Absorption Level d | CYP2D6 Probability e | Num_H Bond Donor | Num_H Bond Acceptor | Molecular Weight | Rotatable Bonds |
|---|---|---|---|---|---|---|---|---|---|
| 6a | 5.21 | 76.04 | 1 | 0 | 0.02 | 1 | 5 | 398.86 | 6 |
| 6b | 5.42 | 76.04 | 1 | 1 | 0 | 1 | 5 | 416.85 | 6 |
| 6c | 5.63 | 76.04 | 1 | 1 | 0 | 1 | 5 | 434.84 | 6 |
| 6d | 5.88 | 76.04 | 1 | 1 | 0.02 | 1 | 5 | 433.31 | 6 |
| 6e | 5.96 | 76.04 | 1 | 1 | 0.02 | 1 | 5 | 477.76 | 6 |
| 6f | 5.11 | 118.86 | 1 | 2 | 0 | 1 | 8 | 443.86 | 7 |
| 8a | 5.14 | 58.74 | 1 | 0 | 0 | 1 | 4 | 384.88 | 6 |
| 8b | 5.80 | 58.74 | 1 | 1 | 0 | 1 | 4 | 419.32 | 6 |
| 8c | 5.88 | 58.74 | 1 | 1 | 0 | 1 | 4 | 463.78 | 6 |
| 9 | 3.20 | 55.25 | 2 | 0 | 0 | 0 | 4 | 349.83 | 5 |
| 10a | 4.51 | 64.71 | 2 | 0 | 0.05 | 1 | 4 | 397.88 | 6 |
| 10b | 5.34 | 64.71 | 2 | 0 | 0 | 1 | 4 | 415.87 | 6 |
| 10c | 5.55 | 64.71 | 1 | 0 | 0 | 1 | 4 | 433.86 | 6 |
| 10d | 5.80 | 64.71 | 1 | 0 | 0.04 | 1 | 4 | 432.32 | 6 |
| 10e | 5.88 | 64.71 | 1 | 0 | 0.02 | 1 | 4 | 476.77 | 6 |
| 10f | 5.03 | 107.54 | 2 | 1 | 0.02 | 1 | 7 | 442.87 | 7 |
| 12a | 5.63 | 51.21 | 1 | 0 | 0 | 0 | 4 | 394.87 | 4 |
| 12b | 5.80 | 51.21 | 1 | 1 | 0 | 0 | 4 | 429.32 | 4 |
| 12c | 6.38 | 51.21 | 1 | 1 | 0 | 0 | 4 | 473.77 | 4 |

3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of (1-(Aryl)-2-(2-(4-fluorophenyl)hydrazono)-2-(phenylsulfonyl)ethanones 6a–f
3.1.2. Synthesis of 1-(Aryl)-2-(1-(4-chlorophenyl)-2-(phenylsulfonyl)ethylidene)hydrazines 8a–c
3.1.3. Synthesis 1-(4-Chlorophenyl)-3-(dimethylamino)-2-(phenylsulfonyl)prop-2-en-1-one (9)
3.1.4. Synthesis of 3-(Aryl amino)-1-(4-chlorophenyl)-2-(phenylsulfonyl)prop-2-en-1-one 10a–f
3.1.5. Synthesis of 1-(4-Aryl)-5-(4-chlorophenyl)-4-(phenylsulfonyl)-1H-pyrazole 12a–c
3.2. Biological Evaluation
3.2.1. Antifungal Activity
3.2.2. In Vitro Cytotoxic Activity
3.3. Docking
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kathiravan, M.K.; Salake, A.B.; Chothe, A.S.; Dudhe, P.B.; Watode, R.P.; Mukta, M.S.; Gadhwe, S. The biology and chemistry of antifungal agents: A review. Bioorg. Med. Chem. 2012, 20, 5678–5698. [Google Scholar] [CrossRef] [PubMed]
- Segura, T.; Puga, A.M.; Burillo, G.; Llovo, J.; Brackman, G.; Coenye, T.; Concheiro, A.; Alvarez-Lorenzo, C. Materials with fungi-bioinspired surface for efficient binding and fungi-sensitive release of antifungal agents. Biomacromolecules 2014, 15, 1860–1870. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, S.; Khosravi, A.R.; Tamai, A.I. Comparative study of microsporum canis isolates by DNA fingerprinting. Mycoses 2014, 57, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Rice, L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Shao, P.L.; Huang, L.M.; Hsueh, P.R. Recent advances and challenges in the treatment of invasive fungal infections. Int. J. Antimicrob. Agents 2007, 30, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Zuber, T.J.; Baddam, K. Superficial fungal infection of the skin. Where and how it appears help determine therapy. Postgrad. Med. 2001, 109, 117–120, 123–126, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in us hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis 2004, 39, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Drake, L.A.; Dinehart, S.M.; Farmer, E.R.; Goltz, R.W.; Graham, G.F.; Hardinsky, M.K.; Lewis, C.W.; Pariser, D.M.; Skouge, J.W.; Webster, S.B.; et al. Guidelines of care for superficial mycotic infections of the skin: Tinea corporis, tinea cruris, tinea faciei, tinea manuum, and tinea pedis. Guidelines/Outcomes Committee. American academy of dermatology. J. Am. Acad. Dermatol. 1996, 34, 282–286. [Google Scholar] [CrossRef]
- Goldstein, A.O.; Smith, K.M.; Ives, T.J.; Goldstein, B. Mycotic infections. Effective management of conditions involving the skin, hair, and nails. Geriatrics 2000, 55, 40–42, 45–47, 51–52. [Google Scholar] [PubMed]
- Lewis, R.E. Current concepts in antifungal pharmacology. Mayo Clin. Proc. 2011, 86, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Cui, C.J.; Chow, E.W.; Pue, N.; Lonhienne, T.; Wang, J.G.; Fraser, J.A.; Guddat, L.W. Sulfonylureas have antifungal activity and are potent inhibitors of candida albicans acetohydroxyacid synthase. J. Med. Chem. 2013, 56, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Clemons, K.V.; Martinez, M.; Calderon, L.; Stevens, D.A. Efficacy of ravuconazole in treatment of systemic murine histoplasmosis. Antimicrob. Agents Chemother. 2002, 46, 922–924. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Boyken, L.; Hollis, R.J.; Kroeger, J.; Messer, S.A.; Tendolkar, S.; Diekema, D.J. In vitro susceptibility of invasive isolates of Candida spp. To anidulafungin, caspofungin, and micafungin: Six years of global surveillance. J. Clin. Microbiol. 2008, 46, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Groll, A.H.; Piscitelli, S.C.; Walsh, T.J. Clinical pharmacology of systemic antifungal agents: A comprehensive review of agents in clinical use, current investigational compounds, and putative targets for antifungal drug development. Adv. Pharmacol. 1998, 44, 343–500. [Google Scholar] [PubMed]
- Kanafani, Z.A.; Perfect, J.R. Antimicrobial resistance: Resistance to antifungal agents: Mechanisms and clinical impact. Clin. Infect. Dis. 2008, 46, 120–128. [Google Scholar] [CrossRef]
- Ashley, E.S.D. Pharmacology of Azole Antifungal Agents; Informa Healthcare USA: New York, NY, USA, 2010. [Google Scholar]
- Yao, B.; Ji, H.; Cao, Y.; Zhou, Y.; Zhu, J.; Lu, J.; Li, Y.; Chen, J.; Zheng, C.; Jiang, Y.; et al. Synthesis and antifungal activities of novel 2-aminotetralin derivatives. J. Med. Chem. 2007, 50, 5293–5300. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.J.; Hitchcock, C.A.; Sibley, C.M. Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 1999, 12, 40–79. [Google Scholar] [PubMed]
- Barone, J.A.; Moskovitz, B.L.; Guarnieri, J.; Hassell, A.E.; Colaizzi, J.L.; Bierman, R.H.; Jessen, L. Food interaction and steady-state pharmacokinetics of itraconazole oral solution in healthy volunteers. Pharmacotherapy 1998, 18, 295–301. [Google Scholar] [PubMed]
- Curti, C.; Laget, M.; Carle, A.O.; Gellis, A.; Vanelle, P. Rapid synthesis of sulfone derivatives as potential anti-infectious agents. Eur. J. Med. Chem. 2007, 42, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Konduru, N.K.; Dey, S.; Sajid, M.; Owais, M.; Ahmed, N. Synthesis and antibacterial and antifungal evaluation of some chalcone based sulfones and bisulfones. Eur. J. Med. Chem. 2013, 59, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Muralikrishna, A.; Venkatesh, B.C.; Padmavathi, V.; Padmaja, A.; Kondaiah, P.; Krishna, N.S. Synthesis, antimicrobial and cytotoxic activities of sulfone linked bis heterocycles. Eur. J. Med. Chem. 2012, 54, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Ghabbour, H.A.; Qabeel, M.M.; Eldehna, W.M.; Al-Dhfyan, A.; Abdel-Aziz, H.A. Design, synthesis, and molecular docking of 1-(1-(4-chlorophenyl)-2-(phenylsulfonyl)ethylidene)-2-phenylhydrazine as potent nonazole anticandidal agent. J. Chem. 2014, 2014, 8. [Google Scholar] [CrossRef]
- Abdel-Aziz, H.A.; Eldehna, W.M.; Fares, M.; Elsaman, T.; Abdel-Aziz, M.M.; Soliman, D.H. Synthesis, in vitro and in silico studies of some novel 5-nitrofuran-2-yl hydrazones as antimicrobial and antitubercular agents. Biol. Pharm. Bull. 2015, 38, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, H.A.; Eldehna, W.M.; Fares, M.; Al-Rashood, S.T.; Al-Rashood, K.A.; Abdel-Aziz, M.M.; Soliman, D.H. Synthesis, biological evaluation and 2D-QSAR study of halophenyl bis-hydrazones as antimicrobial and antitubercular agents. Int. J. Mol. Sci. 2015, 16, 8719–8743. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, H.A.; Mekawey, A.A. Stereoselective synthesis and antimicrobial activity of benzofuran-based (1E)-1-(piperidin-1-yl)-n 2-arylamidrazones. Eur. J. Med. Chem. 2009, 44, 4985–4997. [Google Scholar] [CrossRef] [PubMed]
- Yao, H. Azohydrazone conversion. II. The coupling of diazonium ion with β-diketones. J. Org. Chem. 1964, 29, 2959–2963. [Google Scholar] [CrossRef]
- Abu-Shanab, F.A.; Sherif, S.M.; Mousa, S.A. Dimethylformamide dimethyl acetal as a building block in heterocyclic synthesis. J. Heterocycl. Chem. 2009, 46, 801–827. [Google Scholar] [CrossRef]
- Penning, T.D.; Talley, J.J.; Bertenshaw, S.R.; Carter, J.S.; Collins, P.W.; Docter, S.; Graneto, M.J.; Lee, L.F.; Malecha, J.W.; Miyashiro, J.M. Synthesis and biological evaluation of the 1, 5-diarylpyrazole class of cyclooxygenase-2 inhibitors: Identification of 4-[5-(4-methylphenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl] benzenesulfonamide (SC-58635, celecoxib). J. Med. Chem. 1997, 40, 1347–1365. [Google Scholar] [CrossRef] [PubMed]
- Program, T.D.S. Search for new drugs for treatment of tuberculosis. Antimicrob. Agents Chemother. 2001, 45, 1943–1946. [Google Scholar]
- Podust, L.M.; Poulos, T.L.; Waterman, M.R. Crystal structure of cytochrome p450 14alpha -sterol demethylase (CYP51) from mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 3068–3073. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Fares, M.; Abou-Seri, S.M.; Abdel-Aziz, H.A.; Abbas, S.E.; Youssef, M.M.; Eladwy, R.A. Synthesis and antitumor activity of pyrido [2,3-d]pyrimidine and pyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidine derivatives that induce apoptosis through g1 cell-cycle arrest. Eur. J. Med. Chem. 2014, 83, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not Available.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fares, M.; Said, M.A.; Alsherbiny, M.A.; Eladwy, R.A.; Almahli, H.; Abdel-Aziz, M.M.; Ghabbour, H.A.; Eldehna, W.M.; Abdel-Aziz, H.A. Synthesis, Biological Evaluation and Molecular Docking of Certain Sulfones as Potential Nonazole Antifungal Agents. Molecules 2016, 21, 114. https://doi.org/10.3390/molecules21010114
Fares M, Said MA, Alsherbiny MA, Eladwy RA, Almahli H, Abdel-Aziz MM, Ghabbour HA, Eldehna WM, Abdel-Aziz HA. Synthesis, Biological Evaluation and Molecular Docking of Certain Sulfones as Potential Nonazole Antifungal Agents. Molecules. 2016; 21(1):114. https://doi.org/10.3390/molecules21010114
Chicago/Turabian StyleFares, Mohamed, Mohamed A. Said, Muhammad A. Alsherbiny, Radwa A. Eladwy, Hadia Almahli, Marwa M. Abdel-Aziz, Hazem A. Ghabbour, Wagdy M. Eldehna, and Hatem A. Abdel-Aziz. 2016. "Synthesis, Biological Evaluation and Molecular Docking of Certain Sulfones as Potential Nonazole Antifungal Agents" Molecules 21, no. 1: 114. https://doi.org/10.3390/molecules21010114
APA StyleFares, M., Said, M. A., Alsherbiny, M. A., Eladwy, R. A., Almahli, H., Abdel-Aziz, M. M., Ghabbour, H. A., Eldehna, W. M., & Abdel-Aziz, H. A. (2016). Synthesis, Biological Evaluation and Molecular Docking of Certain Sulfones as Potential Nonazole Antifungal Agents. Molecules, 21(1), 114. https://doi.org/10.3390/molecules21010114