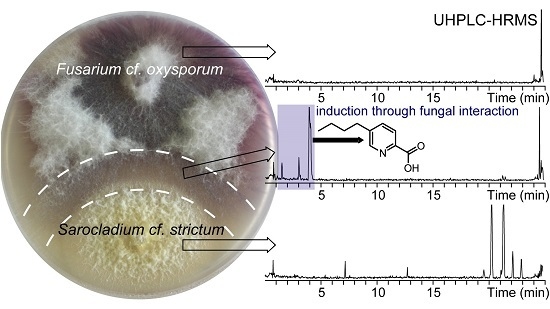
Production of Fusaric Acid by Fusarium spp. in Pure Culture and in Solid Medium Co-Cultures
Abstract
Share and Cite
Bohni, N.; Hofstetter, V.; Gindro, K.; Buyck, B.; Schumpp, O.; Bertrand, S.; Monod, M.; Wolfender, J.-L. Production of Fusaric Acid by Fusarium spp. in Pure Culture and in Solid Medium Co-Cultures. Molecules 2016, 21, 370. https://doi.org/10.3390/molecules21030370
Bohni N, Hofstetter V, Gindro K, Buyck B, Schumpp O, Bertrand S, Monod M, Wolfender J-L. Production of Fusaric Acid by Fusarium spp. in Pure Culture and in Solid Medium Co-Cultures. Molecules. 2016; 21(3):370. https://doi.org/10.3390/molecules21030370
Chicago/Turabian StyleBohni, Nadine, Valérie Hofstetter, Katia Gindro, Bart Buyck, Olivier Schumpp, Samuel Bertrand, Michel Monod, and Jean-Luc Wolfender. 2016. "Production of Fusaric Acid by Fusarium spp. in Pure Culture and in Solid Medium Co-Cultures" Molecules 21, no. 3: 370. https://doi.org/10.3390/molecules21030370
APA StyleBohni, N., Hofstetter, V., Gindro, K., Buyck, B., Schumpp, O., Bertrand, S., Monod, M., & Wolfender, J.-L. (2016). Production of Fusaric Acid by Fusarium spp. in Pure Culture and in Solid Medium Co-Cultures. Molecules, 21(3), 370. https://doi.org/10.3390/molecules21030370