Experimental Evidence and In Silico Identification of Tryptophan Decarboxylase in Citrus Genus
Abstract
:1. Introduction
2. Results and Discussion
2.1. TDC Activity and Identification of Labeled Tryptamine and Its Labeled Derivative in Citrus Seedlings
2.2. In Silico Identification and Annotation of Putative TDC Sequences in Citrus
2.2.1. Sequence Similarity to the C. roseus TDC
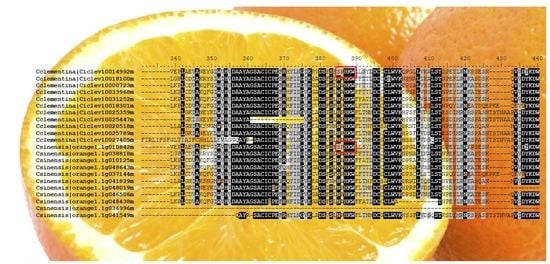
2.2.2. Multiple Sequence Alignments and Identification of Specific TDC Signatures
2.2.3. Citrus Putative TDC Sequence Analysis and Annotation Improvement
3. Materials and Methods
3.1. Reagents
3.2. TDC Activity Assay
3.3. Preparation of Seedlings
3.4. Analysis of Tryptamine and Its N-methyl Derivatives by HPLC-ESI-MS/MS
3.5. Bioinformatics Analyses
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- De Luca, V.; Marineau, C.; Brisson, N. Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: comparison with animal dopa decarboxylases. Proc. Natl. Acad. Sci. USA 1989, 86, 2582–2586. [Google Scholar] [CrossRef] [PubMed]
- Sandmeier, E.; Hale, T.I.; Christen, P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 1994, 221, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Servillo, L.; Giovane, A.; Balestrieri, M.L.; Casale, R.; Cautela, D.; Castaldo, D. Citrus genus plants contain N-methylated tryptamine derivatives and their 5-hydroxylated forms. J. Agric. Food. Chem. 2013, 61, 5156–5162. [Google Scholar] [CrossRef] [PubMed]
- Bartley, G.E.; Breksa, A.P., III; Ishida, B.K. PCR amplification and cloning of tyrosine decarboxylase involved in synephrine biosynthesis in Citrus. New Biotechnol. 2010, 27, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Servillo, L.; Giovane, A.; D’Onofrio, N.; Casale, R.; Cautela, D.; Ferrari, G.; Balestrieri, M.L.; Castaldo, D. N-methylated derivatives of tyramine in Citrus genus plants: identification of N,N,N-trimethyltyramine (candicine). J. Agric. Food Chem. 2014, 62, 2679–2684. [Google Scholar] [CrossRef] [PubMed]
- Goddijn, O.J.; Lohman, F.P.; de Kam, R.J.; Schilperoort, R.A.; Hoge, J.H. Nucleotide sequence of the tryptophan decarboxylase gene of Catharanthus roseus and expression of tdc-gusA gene fusions in Nicotiana tabacum. Mol. Gen. Genet. 1994, 242, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Guillet, G.; Poupart, J.; Basurco, J.; de Luca, V. Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiol. 2000, 122, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J.; Huber-Allanach, K.L.; Tari, L.W. Plant aromatic l-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 2000, 54, 121–138. [Google Scholar] [CrossRef]
- Sajem, A.L.; Gosai, K. Traditional use of medicinal plants by the Jaintia tribes in North Cachar Hills district of Assam, northeast India. J. Ethnobiol. Ethnomed. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J.; De Luca, V. Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 2008, 54, 763–784. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.I.; Kennedy, T.J.; Christian, D.R., Jr. Tryptamines. In Basic Principles of Forensic Chemistry; Humana Press: New York, NY, USA, 2012; pp. 191–206. [Google Scholar]
- Servillo, L.; Giovane, A.; Balestrieri, M.L.; Cautela, D.; Castaldo, D. N-methylated tryptamine derivatives in Citrus genus plants: identification of N,N,N-trimethyltryptamine in bergamot. J. Agric. Food Chem. 2012, 60, 9512–9518. [Google Scholar] [CrossRef] [PubMed]
- Servillo, L.; Giovane, A.; Casale, R.; D’Onofrio, N.; Ferrari, F.; Cautela, D.; Balestrieri, M.L.; Castaldo, D. Serotonin 5-O-β-glucoside and its N-methylated forms in Citrus genus plants. J. Agric. Food Chem. 2015, 63, 4220–4227. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C.; Adams, D.C.; Nessler, C.L.; Brown, J.K.; Bohnert, H.J. Tryptophan decarboxylase, tryptamine, and reproduction of the whitefly. Plant Physiol. 1995, 109, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.I.; Ellis, B.E.; Isman, M.B. Tryptamine-induced resistance in tryptophan decarboxylase transgenic poplar and tobacco plants against their specific herbivores. J. Chem. Ecol. 2003, 29, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, A.; Nakao, T.; Mashimo, Y.; Murai, M.; Ichimaru, N.; Tanaka, C.; Nakajima, H.; Wakasa, K.; Miyagawa, H. Probing the role of tryptophan-derived secondary metabolism in defense responses against Bipolaris oryzae infection in rice leaves by a suicide substrate of tryptophan decarboxylase. Phytochemistry 2011, 72, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Pignone, D.; Hammer, K. Conservation, evaluation, and utilization of biodiversity. In Genomics and Breeding for Climate-Resilient Crops; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 1, pp. 9–26. [Google Scholar]
- Esposito, C.; De Masi, L.; Laratta, B.; Tridente, R.L.; Castaldo, D. Sintesi di limonoidi appartenenti al pathway biosintetico della limonina e dei 7α-acetilati. Essenze Derivati Agrumari 2005, 75, 83–94. [Google Scholar]
- Esposito, C.; Tridente, R.L.; Balestrieri, M.L.; Laratta, B.; De Masi, L.; Servillo, L.; Castaldo, D. Distribuzione dei limonoidi nelle diverse parti del frutto di bergamotto. Essenze Derivati Agrumari 2006, 76, 11–18. [Google Scholar]
- Laratta, B.; De Masi, L.; Minasi, P.; Giovane, A. Pectin methylesterase in Citrus bergamia R.: purification, biochemical characterization and sequence of the exon related to the enzyme active site. Food Chem. 2008, 110, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Servillo, L.; D’Onofrio, N.; Longobardi, L.; Sirangelo, I.; Giovane, A.; Cautela, D.; Castaldo, D.; Giordano, A.; Balestrieri, M.L. Stachydrine ameliorates high-glucose induced endothelial cell senescence and SIRT1 downregulation. J. Cell. Biochem. 2013, 114, 2522–2530. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; Rokhsar, D.S. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J. Alkaloid biosynthesis in plants: Biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 29–66. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.K.; Menezes-Benavente, L.; Galvão, V.C.; Margis-Pinheiro, M. Multigene families encode the major enzymes of antioxidant metabolism in Eucalyptus grandis L. Genet. Mol. Biol. 2005, 28 (Suppl.), 529–538. [Google Scholar] [CrossRef]
- Facchini, P.J.; Bird, D.A.; St-Pierre, B. Can Arabidopsis make complex alkaloids? TRENDS Plant Sci. 2004, 9, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Mizuguchi, H.; Nishino, J.; Hayashi, H.; Kagamiyama, H. Functionally important residues of aromatic l-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J. Biochem. 1996, 120, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, P.; Dominici, P.; Borri-Voltattorni, C.; Jansonius, J.N.; Malashkevich, V.N. Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol. 2001, 8, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Ding, H.; Robinson, H.; Christensen, B.M.; Li, J. Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase. PLoS ONE 2010, 5, e8826. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kang, K.; Lee, K.; Choi, D.; Kim, Y.S.; Back, K. Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in pepper fruits (Capsicum annuum) upon pathogen infection. Planta 2009, 230, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.


| Plant Species | GenBank Accession No. |
|---|---|
| TDC (14 Sequences) | |
| Actaea racemosa | ADD71923, ADD71924, CCO62221 |
| Camptotheca acuminata | AAB39708, AAB39709 |
| Capsicum annuum | ACN62126, ACN62127 |
| Catharanthus roseus | P17770 |
| Mitragyna speciosa | AEQ01059 |
| Ophiorrhiza prostrata | ABU40982 |
| Ophiorrhiza pumila | BAC41515 |
| Rauvolfia verticillata | ADL28270 |
| Tabernaemontana elegans | AEY82396 |
| Vinca minor | AEY82397 |
| TYDC (28 Sequences) | |
| Arabidopsis thaliana | NP_001078461, NP_194597, CAB56038, NP_849999 |
| Argemone mexicana | ACJ76782 |
| Aristolochia contorta | ABJ16446 |
| Citrus aurantium | ACX29995 |
| Citrus reshni | ACX29990, ACX29991 |
| Citrus reticulata | ACX29996 |
| Citrus sinensis | ACX29992 |
| Medicago truncatula | AES81613, AES81615 |
| Papaver somniferum | from P54768 to P54771 |
| from AAC61841 to AAC61844 | |
| Petroselinum crispum | from Q06085 to Q06088 |
| Solanum tuberosum | AHI16967 |
| Thalictrum flavum | AAG60665 |
| Theobroma cacao | EOX96928 |
| TDC Motif (Position) | TYDC Motif (Position) | Hypothetical Binding 1 |
|---|---|---|
| T[H/N]W[L/M]SP (92) | THWQSP (137) | Substrate |
| FPATVSSAAF (103) | [F/Y][P/A]S[S/N][G/S/T]S[I/V/T]AGF (148) | Substrate |
| [H/Q][G/N]TTSE[A/S]ILCT(167) | QGT[T/A/S][C/S]EA[V/I]L[C/V][T/V] (217) | Substrate |
| SPHKW (318) | NAHKW (369) | Substrate, PLP |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Masi, L.; Castaldo, D.; Pignone, D.; Servillo, L.; Facchiano, A. Experimental Evidence and In Silico Identification of Tryptophan Decarboxylase in Citrus Genus. Molecules 2017, 22, 272. https://doi.org/10.3390/molecules22020272
De Masi L, Castaldo D, Pignone D, Servillo L, Facchiano A. Experimental Evidence and In Silico Identification of Tryptophan Decarboxylase in Citrus Genus. Molecules. 2017; 22(2):272. https://doi.org/10.3390/molecules22020272
Chicago/Turabian StyleDe Masi, Luigi, Domenico Castaldo, Domenico Pignone, Luigi Servillo, and Angelo Facchiano. 2017. "Experimental Evidence and In Silico Identification of Tryptophan Decarboxylase in Citrus Genus" Molecules 22, no. 2: 272. https://doi.org/10.3390/molecules22020272
APA StyleDe Masi, L., Castaldo, D., Pignone, D., Servillo, L., & Facchiano, A. (2017). Experimental Evidence and In Silico Identification of Tryptophan Decarboxylase in Citrus Genus. Molecules, 22(2), 272. https://doi.org/10.3390/molecules22020272