Figure 1.
Structure of glycyrrhetinic acid.
Figure 1.
Structure of glycyrrhetinic acid.
Scheme 1.
Synthesis of the GA amino alkyl derivatives 1–11. Reagents and conditions: (a) K2CO3,CH3I, DMF, 24 h, 25 °C; (b) ClCH2COCl, Et3N, THF (or CH2Cl2), 25 °C, 12 h; (c) H2N-(CH2)n-NH2, DMF, K2CO3, 12 h, 25 °C.
Scheme 1.
Synthesis of the GA amino alkyl derivatives 1–11. Reagents and conditions: (a) K2CO3,CH3I, DMF, 24 h, 25 °C; (b) ClCH2COCl, Et3N, THF (or CH2Cl2), 25 °C, 12 h; (c) H2N-(CH2)n-NH2, DMF, K2CO3, 12 h, 25 °C.
Scheme 2.
Synthesis of the GA amino acid derivatives 12–32. Reagents and conditions: (a) K2CO3, alkyl halides, DMF, 24 h, 25 °C; (b) These compounds were synthesized by DCC mediated esterification of N-Boc protected amino acids followed by their deportation using TFA in dry DCM (for the amines) or by treating them with dry HCl gas in DCM (for the ammonium hydrochlorides).
Scheme 2.
Synthesis of the GA amino acid derivatives 12–32. Reagents and conditions: (a) K2CO3, alkyl halides, DMF, 24 h, 25 °C; (b) These compounds were synthesized by DCC mediated esterification of N-Boc protected amino acids followed by their deportation using TFA in dry DCM (for the amines) or by treating them with dry HCl gas in DCM (for the ammonium hydrochlorides).
Scheme 3.
Synthesis of the GA amino acid derivatives 33–44. Reagents and conditions: (a) DCC, DMAP, Boc-Asp(OBzl)OH or Boc-Glu(OBzl)OH, DCM, 12 h, 25 °C; (b) TFA, DCM, 12 h, 25 °C; (c) NH4+HCO2−, Pd/C (10%), THF/MeOH, 12 h, 25 °C; (d) HCl (gas), DCM, 12 h, 25 °C; (e) CH3I, K2CO3, DMF, 2 h, 25 °C.
Scheme 3.
Synthesis of the GA amino acid derivatives 33–44. Reagents and conditions: (a) DCC, DMAP, Boc-Asp(OBzl)OH or Boc-Glu(OBzl)OH, DCM, 12 h, 25 °C; (b) TFA, DCM, 12 h, 25 °C; (c) NH4+HCO2−, Pd/C (10%), THF/MeOH, 12 h, 25 °C; (d) HCl (gas), DCM, 12 h, 25 °C; (e) CH3I, K2CO3, DMF, 2 h, 25 °C.
Scheme 4.
Synthesis of the GA–Me (GA methyl ester) amino ester derivatives 45–59. Reagents and conditions: (a) Boc-amino acids, DCM, DMAP, DCC, 12 h, 25 °C; (b) TFA in DCM, 12 h, 25 °C, or HCl (gas) in DCM, 12 h, 25 °C.
Scheme 4.
Synthesis of the GA–Me (GA methyl ester) amino ester derivatives 45–59. Reagents and conditions: (a) Boc-amino acids, DCM, DMAP, DCC, 12 h, 25 °C; (b) TFA in DCM, 12 h, 25 °C, or HCl (gas) in DCM, 12 h, 25 °C.
Scheme 5.
Synthesis of the GA glycosides derivatives 60–66. Reagents and conditions: (a) Sugar trichloro acetimidate, TMSOTf, DCM, −70 °C–25 °C, 2 h.
Scheme 5.
Synthesis of the GA glycosides derivatives 60–66. Reagents and conditions: (a) Sugar trichloro acetimidate, TMSOTf, DCM, −70 °C–25 °C, 2 h.
Scheme 6.
Synthesis of the GA furan-based nitric oxide (NO)-releasing derivatives 67–74. Reagents and conditions: (a) CH3OH, p-TSA; (b) succinic anhydride, DMAP, dry DCM, 15 h; (c) phenylsulfonyl furans, DCC, DMAP, dry DCM, 24 h.
Scheme 6.
Synthesis of the GA furan-based nitric oxide (NO)-releasing derivatives 67–74. Reagents and conditions: (a) CH3OH, p-TSA; (b) succinic anhydride, DMAP, dry DCM, 15 h; (c) phenylsulfonyl furans, DCC, DMAP, dry DCM, 24 h.
Figure 2.
Structures of GA-1, GA-2 and GA-3.
Figure 2.
Structures of GA-1, GA-2 and GA-3.
Figure 3.
Structures of CDDO, CDDO-Me, β-CDODA and β-CDODA-Me.
Figure 3.
Structures of CDDO, CDDO-Me, β-CDODA and β-CDODA-Me.
Scheme 7.
Synthesis of the GA 2-substituted derivatives 75–79. Reagents and conditions: (a) CH2N2, Et2O, 0 °C; (b) IBX, DMSO, 21 h, 80–85 °C; (c) iodine, pyridine, tetrahydrofuran; (d) CuCN, NMP, 2 h, 130 °C; (e) CH3SO2Na, CuI, DMSO, 20 h, 120–125 °C; (f) CuI, methyl-2,2-difluoro-2-(fluorosulfonyl) acetate, DMF/HMPT, 20 h, 70 °C; (g) dimethyl phosphite, Cs2CO3, N,N-dimethylethylenediamine, toluene, 26 h, 95–100 °C.
Scheme 7.
Synthesis of the GA 2-substituted derivatives 75–79. Reagents and conditions: (a) CH2N2, Et2O, 0 °C; (b) IBX, DMSO, 21 h, 80–85 °C; (c) iodine, pyridine, tetrahydrofuran; (d) CuCN, NMP, 2 h, 130 °C; (e) CH3SO2Na, CuI, DMSO, 20 h, 120–125 °C; (f) CuI, methyl-2,2-difluoro-2-(fluorosulfonyl) acetate, DMF/HMPT, 20 h, 70 °C; (g) dimethyl phosphite, Cs2CO3, N,N-dimethylethylenediamine, toluene, 26 h, 95–100 °C.
Scheme 8.
Synthesis of the C-2 and C-3 modified GA derivatives 80–97. Reagents and conditions: (a) AcCl, pyridine, CH2Cl2, 2 h, 25 °C; (b) Jones reagent, 20–60 min, 25 °C; (c) KOH, hydrazine, ethylene glycol, 24 h, 200 °C; (d) periodic acid, DMSO, 3 days, −50 °C; (e) HOAc, p-TsOH, 24 h, 80 °C; (f) MeSO2Cl, pyridine (or Et3N for 15), 1–70 h, 25 °C; (g) for 92: K2CO3, DMF, 24 h, 120 °C; for 93: Bu4NF, DMF, 4 days, 102 °C; for 94: PPh3, 3,3-dimethylglutarimide, DEAD, THF, 24 h, 25 °C; (h) m-CPBA, CH2Cl2, 20 h, 25 °C; (i) 1,1′-thiocarbonyldiimidazole, 1,2-dichloroethane, 70 h, 100 °C; (j) Bu3SnH, AIBN (cat.), toluene, 40 h, 115 °C.
Scheme 8.
Synthesis of the C-2 and C-3 modified GA derivatives 80–97. Reagents and conditions: (a) AcCl, pyridine, CH2Cl2, 2 h, 25 °C; (b) Jones reagent, 20–60 min, 25 °C; (c) KOH, hydrazine, ethylene glycol, 24 h, 200 °C; (d) periodic acid, DMSO, 3 days, −50 °C; (e) HOAc, p-TsOH, 24 h, 80 °C; (f) MeSO2Cl, pyridine (or Et3N for 15), 1–70 h, 25 °C; (g) for 92: K2CO3, DMF, 24 h, 120 °C; for 93: Bu4NF, DMF, 4 days, 102 °C; for 94: PPh3, 3,3-dimethylglutarimide, DEAD, THF, 24 h, 25 °C; (h) m-CPBA, CH2Cl2, 20 h, 25 °C; (i) 1,1′-thiocarbonyldiimidazole, 1,2-dichloroethane, 70 h, 100 °C; (j) Bu3SnH, AIBN (cat.), toluene, 40 h, 115 °C.
Scheme 9.
Synthesis of ring A modified GA derivatives 98–112. Reagents and conditions: (a) Jones’ reagent; (b) HCO2Et, NaOMe; (c) NaOMe, H2O2; (d) t-BuOK/t-BuOH, n-BuONO; (e) NaBH4; (f) p-TsCl; (g) CH3I, K2CO3; (h) LiBr, Li2CO3; (i) m-CPBA, K2CO3; (j) HClO4; (k) KOH; (l) m-CPBA, NaHCO3; (m) NaOMe; (n) NH2OH·HCl; (o) p-TsCl, DMAP.
Scheme 9.
Synthesis of ring A modified GA derivatives 98–112. Reagents and conditions: (a) Jones’ reagent; (b) HCO2Et, NaOMe; (c) NaOMe, H2O2; (d) t-BuOK/t-BuOH, n-BuONO; (e) NaBH4; (f) p-TsCl; (g) CH3I, K2CO3; (h) LiBr, Li2CO3; (i) m-CPBA, K2CO3; (j) HClO4; (k) KOH; (l) m-CPBA, NaHCO3; (m) NaOMe; (n) NH2OH·HCl; (o) p-TsCl, DMAP.
Scheme 10.
Synthesis of ring C modified GA derivatives 113–115. Reagents and conditions: (a) Zinc dust, conc. HCl, dioxane, 25 °C, 24 h; (b) MeI, K2CO3, DMF, 25 °C, 24 h; (c) BH3-THF, THF, citric acid, 25 °C, 20 h; (d) EtI, K2CO3, DMF, 25 °C, 24 h; (e) BH3-THF, THF, Na2CO3, 25 °C, 4 days.
Scheme 10.
Synthesis of ring C modified GA derivatives 113–115. Reagents and conditions: (a) Zinc dust, conc. HCl, dioxane, 25 °C, 24 h; (b) MeI, K2CO3, DMF, 25 °C, 24 h; (c) BH3-THF, THF, citric acid, 25 °C, 20 h; (d) EtI, K2CO3, DMF, 25 °C, 24 h; (e) BH3-THF, THF, Na2CO3, 25 °C, 4 days.
Scheme 11.
Synthesis of ring E modified GA derivatives 116–130. Reagents and conditions: (a) 1. DCC, HOBt, DIPEA, DMF, r.t., 30 min; 2. R1NH2, r.t., overnight; (b) 1. DCC, HOBt, DIPEA, DMF, r.t., 30 min; 2. H2N(CH2)2NHBoc, r.t., overnight; (c) TFA, DCM, 0 °C, 3 h; (d) DMAP, RCOCl, DCM; (e) THF, RNCO, r.t., 20 h; (f) THF, RNCS, r.t., 20 h; (g) Jones reagent, acetone, 0 °C, 45 min.
Scheme 11.
Synthesis of ring E modified GA derivatives 116–130. Reagents and conditions: (a) 1. DCC, HOBt, DIPEA, DMF, r.t., 30 min; 2. R1NH2, r.t., overnight; (b) 1. DCC, HOBt, DIPEA, DMF, r.t., 30 min; 2. H2N(CH2)2NHBoc, r.t., overnight; (c) TFA, DCM, 0 °C, 3 h; (d) DMAP, RCOCl, DCM; (e) THF, RNCO, r.t., 20 h; (f) THF, RNCS, r.t., 20 h; (g) Jones reagent, acetone, 0 °C, 45 min.
Scheme 12.
Synthesis of ring E modified GA derivatives 131–155. Reagents and conditions: (a) (i) amino acid methyl ester EDCI/HOBt/Et3N, DMF; (ii) 4N NaOH THF/MeOH; (b) EDCI/HOBt/Et3N DMF, r.t., overnight; (c) EDCI/HOBt/Et3N DMF, r.t., overnight.
Scheme 12.
Synthesis of ring E modified GA derivatives 131–155. Reagents and conditions: (a) (i) amino acid methyl ester EDCI/HOBt/Et3N, DMF; (ii) 4N NaOH THF/MeOH; (b) EDCI/HOBt/Et3N DMF, r.t., overnight; (c) EDCI/HOBt/Et3N DMF, r.t., overnight.
Scheme 13.
Syntheses of GA–DZ derivatives
156–
166.
Reagents and conditions: (a)
![Molecules 22 00924 i001]()
, 1N NaOH (for R
2 = Me), 5N KOH (for R
2 = Ph); (b) GA, EDCI, DMAP, CH
2Cl
2.
Scheme 13.
Syntheses of GA–DZ derivatives
156–
166.
Reagents and conditions: (a)
![Molecules 22 00924 i001]()
, 1N NaOH (for R
2 = Me), 5N KOH (for R
2 = Ph); (b) GA, EDCI, DMAP, CH
2Cl
2.
Scheme 14.
Synthesis of the GA amide derivatives 167–169. Reagents and conditions: (a) K2CO3, diamine, DMF, 25 °C, 20 h; (b) Boc2O, Et3N, MeOH, 25 °C, 20 h.
Scheme 14.
Synthesis of the GA amide derivatives 167–169. Reagents and conditions: (a) K2CO3, diamine, DMF, 25 °C, 20 h; (b) Boc2O, Et3N, MeOH, 25 °C, 20 h.
Scheme 15.
Synthesis of the GA ester derivatives 170–175. Reagents and conditions: (a) K2CO3, alkyl halide, DMF, 25 °C, 20 h.
Scheme 15.
Synthesis of the GA ester derivatives 170–175. Reagents and conditions: (a) K2CO3, alkyl halide, DMF, 25 °C, 20 h.
Scheme 16.
Synthesis of multiple rings modified GA derivatives 176–199. Reagents and conditions: (a) ferulic acid analogs, EDCI, DMAP, CH2Cl2, r.t.; (b) Boc-L-methionine or Boc-L-selenomethionine, EDCI, DMAP, CH2Cl2, r.t.; (c) HCl (gas) in CH2Cl2, r.t.
Scheme 16.
Synthesis of multiple rings modified GA derivatives 176–199. Reagents and conditions: (a) ferulic acid analogs, EDCI, DMAP, CH2Cl2, r.t.; (b) Boc-L-methionine or Boc-L-selenomethionine, EDCI, DMAP, CH2Cl2, r.t.; (c) HCl (gas) in CH2Cl2, r.t.
Scheme 17.
Synthesis of multiple rings modified GA derivatives 200–210: Reagents and conditions: (a) K2CO3, cat. KI, 60 °C, 12 h, chromatography; (b) Ac2O, Py, r.t., 3 h, chromatography; (c) K2CO3, cat. KI, 224, or 225 or 226, 60 °C, 12 h; (d) Zn (containing 10% HgCl2), concentrated. HCl, 1,4-dioxane, 20 °C, 2 h, chromatography; (e) K2CO3, cat. KI, 227, 60 °C, 12 h, chromatography; (f) ClCH2COCl, Py, THF, r.t., 4 h; (g) Et3N, THF, refluxing, 10 h; (h) K2CO3, cat. KI 230 or 231 60 °C, 12 h, chromatography.
Scheme 17.
Synthesis of multiple rings modified GA derivatives 200–210: Reagents and conditions: (a) K2CO3, cat. KI, 60 °C, 12 h, chromatography; (b) Ac2O, Py, r.t., 3 h, chromatography; (c) K2CO3, cat. KI, 224, or 225 or 226, 60 °C, 12 h; (d) Zn (containing 10% HgCl2), concentrated. HCl, 1,4-dioxane, 20 °C, 2 h, chromatography; (e) K2CO3, cat. KI, 227, 60 °C, 12 h, chromatography; (f) ClCH2COCl, Py, THF, r.t., 4 h; (g) Et3N, THF, refluxing, 10 h; (h) K2CO3, cat. KI 230 or 231 60 °C, 12 h, chromatography.
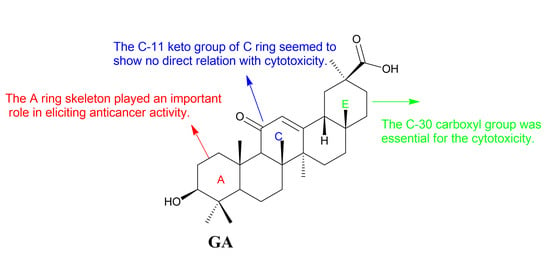
Figure 4.
Structure-activity relationships of GA.
Figure 4.
Structure-activity relationships of GA.
Table 1.
Cytotoxicity (IC50 values in µM) of 1–11 in a panel of various cancer cell lines.
Table 1.
Cytotoxicity (IC50 values in µM) of 1–11 in a panel of various cancer cell lines.
| Cell Lines | GA | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|
| 518A2 | 83.92 | 27.54 | 25.43 | 5.24 | 3.79 | 2.55 | 2.02 | 1.09 | 1.27 | 3.49 | 3.12 | 4.33 |
| 8505C | 86.50 | 26.07 | 26.08 | 15.86 | 3.37 | 2.12 | 1.78 | 1.68 | 2.13 | 3.35 | 6.18 | 7.60 |
| A253 | 80.78 | 19.42 | 25.54 | 6.19 | 3.64 | 2.56 | 2.27 | 1.12 | 1.74 | 3.01 | 4.65 | 5.48 |
| A2780 | 74.57 | 25.54 | 23.77 | 6.01 | 4.39 | 2.43 | 2.00 | 1.36 | 1.14 | 2.80 | 3.30 | 3.63 |
| A549 | 82.76 | 23.50 | 24.80 | 8.39 | 5.15 | 3.31 | 2.52 | 1.59 | 2.21 | 4.08 | 2.23 | 5.16 |
| DLD-1 | 81.21 | 26.12 | 17.36 | 6.13 | 4.39 | 2.66 | 2.40 | 0.91 | 1.25 | 3.96 | 4.50 | 5.53 |
| FADU | 84.55 | 23.41 | 23.56 | 12.44 | 5.57 | 3.51 | 3.30 | 1.78 | 2.20 | 4.26 | 5.54 | 5.65 |
| HCT-11 | 78.83 | 22.10 | 14.41 | 5.13 | 4.30 | 2.41 | 2.19 | 1.17 | 1.70 | 3.53 | 3.44 | 3.86 |
| HCT-8 | 78.85 | 24.36 | 13.39 | 3.97 | 2.37 | 1.51 | 1.38 | 0.62 | 0.89 | 2.92 | 2.42 | 4.07 |
| HT-29 | 80.09 | 27.54 | 16.91 | 5.34 | 2.90 | 1.69 | 1.28 | 0.59 | 0.86 | 2.76 | 2.06 | 2.73 |
| LIPO | 81.44 | 20.47 | 25.39 | 14.55 | 3.89 | 2.57 | 1.93 | 1.59 | 1.44 | 4.36 | 5.48 | 6.93 |
| MCF-7 | 84.70 | 22.14 | 25.22 | 6.69 | 3.55 | 2.45 | 1.79 | 1.17 | 0.98 | 3.89 | 3.33 | 2.68 |
| SW1736 | 76.93 | 34.87 | 16.42 | 3.14 | 6.05 | 3.30 | 2.69 | 1.61 | 2.24 | 4.09 | 3.30 | 3.73 |
| SW480 | 86.80 | 16.08 | 25.91 | 8.92 | 3.68 | 2.54 | 1.91 | 2.25 | 2.24 | 3.93 | 5.74 | 4.73 |
Table 2.
Cytotoxicity (IC50 values in µM) of 12–32 in a panel of various cancer cell lines.
Table 2.
Cytotoxicity (IC50 values in µM) of 12–32 in a panel of various cancer cell lines.
| Compound | 8505C | A253 | A2780 | A549 | DLD-1 | LIPO | Average |
|---|
| GA | 86.50 ± 4.20 | 80.78 ± 4.04 | 74.57 ± 3.73 | 82.76 ± 4.14 | 81.21 ± 4.06 | 81.44 ± 4.07 | 81.4 ± 4.07 |
| 12 | 24.58 ± 1.23 | 25.04 ± 1.25 | 26.96 ± 1.35 | 22.74 ± 1.14 | 28.14 ± 1.41 | 27.66 ± 1.38 | 24.39 ± 1.22 |
| 13 | 14.24 ± 0.71 | 15.76 ± 0.79 | 24.95 ± 1.25 | 14.41 ± 0.72 | 27.61 ± 1.38 | 15.93 ± 0.80 | 19.21 ± 0.96 |
| 14 | 8.10 ± 0.41 | 10.67 ± 0.54 | 20.32 ± 1.18 | 6.15 ± 0.31 | 22.69 ± 1.13 | 11.54 ± 0.80 | 13.76 ± 0.69 |
| 15 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 16 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 17 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 18 | 7.45 ± 0.37 | 6.26 ± 0.31 | 5.99 ± 0.30 | 6.42 ± 0.32 | 8.59 ± 0.43 | 7.54 ± 0.38 | 7.04 ± 0.35 |
| 19 | 4.31 ± 0.22 | 3.61 ± 0.18 | 2.98 ± 0.15 | 2.77 ± 0.14 | 4.49 ± 0.22 | 4.30 ± 0.22 | 3.74 ± 0.19 |
| 20 | 2.55 ± 0.13 | 2.50 ± 0.13 | 1.72 ± 0.09 | 2.40 ± 0.12 | 2.51 ± 0.13 | 2.52 ± 0.13 | 2.37 ± 0.12 |
| 21 | 5.32 ± 0.27 | 3.59 ± 0.18 | 3.90 ± 0.20 | 5.39 ± 0.27 | 5.61 ± 0.28 | 4.32 ± 0.22 | 4.69 ± 0.23 |
| 22 | 3.87 ± 0.19 | 2.33 ± 0.12 | 2.59 ± 0.13 | 3.43 ± 0.17 | 3.72 ± 0.19 | 2.74 ± 0.14 | 3.11 ± 0.16 |
| 23 | 2.32 ± 0.12 | 2.23 ± 0.11 | 1.77 ± 0.09 | 2.18 ± 0.11 | 2.74 ± 0.14 | 2.38 ± 0.12 | 2.27 ± 0.11 |
| 24 | 2.76 ± 0.14 | 2.01 ± 0.10 | 2.24 ± 0.11 | 2.65 ± 0.13 | 2.54 ± 0.13 | 2.74 ± 0.14 | 2.49 ± 0.12 |
| 25 | 3.49 ± 0.17 | 3.51 ± 0.18 | 2.08 ± 0.10 | 3.43 ± 0.17 | 5.54 ± 0.28 | 3.53 ± 0.18 | 3.60 ± 0.18 |
| 26 | 1.96 ± 0.10 | 2.68 ± 0.13 | 1.31 ± 0.07 | 1.78 ± 0.09 | 3.52 ± 0.18 | 3.49 ± 0.17 | 2.46 ± 0.12 |
| 27 | 4.79 ± 0.24 | 5.03 ± 0.25 | 3.54 ± 0.18 | 5.07 ± 0.25 | 4.54 ± 0.23 | 4.81 ± 0.24 | 4.63 ± 0.23 |
| 28 | 3.10 ± 0.16 | 3.49 ± 0.17 | 2.85 ± 0.14 | 3.51 ± 0.18 | 5.02 ± 0.25 | 3.57 ± 0.18 | 3.59 ± 0.18 |
| 29 | 3.19 ± 0.16 | 3.05 ± 0.15 | 1.73 ± 0.09 | 2.76 ± 0.14 | 4.54 ± 0.23 | 3.25 ± 0.16 | 3.09 ± 0.15 |
| 30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 31 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 32 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
Table 3.
Cytotoxicity (IC50 values in µM) of 33–40, 43, 44 in a panel of various cancer cell lines.
Table 3.
Cytotoxicity (IC50 values in µM) of 33–40, 43, 44 in a panel of various cancer cell lines.
| Cell Lines | 35 | 36 | 37 | 38 | 39 | 40 | 43 | 44 |
|---|
| 518A2 | 10.90 ± 0.55 | 1.75 ± 0.09 | 17.19 ± 0.86 | 17.94 ± 0.90 | >100 | >100 | 39.24 ± 1.96 | 47.72 ± 2.39 |
| 8505C | 12.97 ± 0.45 | 1.76 ± 0.09 | 15.82 ± 0.79 | 17.00 ± 0.85 | >100 | >100 | 45.36 ± 2.27 | 61.57 ± 3.08 |
| A253 | 7.99 ± 0.40 | 1.28 ± 0.06 | 15.07 ± 0.75 | 13.80 ± 0.69 | >100 | >100 | 30.47 ± 1.52 | 53.07 ± 2.65 |
| A2780 | 8.84 ± 0.44 | 1.65 ± 0.08 | 17.29 ± 0.86 | 18.24 ± 0.91 | >100 | >100 | 22.44 ± 1.12 | 29.19 ± 1.46 |
| A549 | 10.94 ± 0.55 | 1.77 ± 0.09 | 19.82 ± 0.99 | 21.20 ± 1.06 | >100 | >100 | 31.59 ± 1.58 | 60.96 ± 3.05 |
| Lipo | 11.35 ± 0.57 | 1.74 ± 0.09 | 16.67 ± 0.83 | 18.78 ± 0.94 | >100 | >100 | 40.62 ± 2.03 | 54.77 ± 2.74 |
| MCF-7 | 7.35 ± 0.36 | 1.27 ± 0.06 | 17.47 ± 0.87 | 16.96 ± 0.85 | >100 | >100 | 16.89 ± 0.84 | 29.26 ± 1.46 |
| SW1736 | 16.68 ± 0.83 | 2.33 ± 0.12 | 17.13 ± 0.86 | 19.24 ± 0.96 | >100 | >100 | 20.85 ± 1.04 | 38.50 ± 1.93 |
| Average | 10.88 ± 0.54 | 1.69 ± 0.08 | 17.06 ± 0.85 | 17.90 ± 0.90 | >100 | >100 | 30.93 ± 1.55 | 46.77 ± 2.34 |
| NiH3T3 | 14.74 ± 0.74 | 39.09 ± 1.95 | 23.09 ± 1.15 | 24.42 ± 1.22 | >100 | >100 | 16.89 ± 0.84 | 33.63 ± 1.68 |
| F | 1.35 | 23.13 | 1.35 | 1.36 | | | 0.55 | 0.72 |
Table 4.
Cytotoxicity (IC50 values in µM) of 45–59 in a panel of various cancer cell lines.
Table 4.
Cytotoxicity (IC50 values in µM) of 45–59 in a panel of various cancer cell lines.
| Compound | 8505C | A253 | A2780 | A549 | DLD-1 | LIPO | MCF-7 |
|---|
| 45 | 2.92 | 2.26 | 2.24 | 2.26 | 3.35 | 3.56 | 2.25 |
| 46 | 2.50 | 2.46 | 1.83 | 2.13 | 3.42 | 2.50 | 2.49 |
| 47 | 9.62 | 5.56 | 4.58 | 6.91 | 11.64 | 7.96 | 5.49 |
| 48 | 16.93 | 6.41 | 5.50 | 9.94 | 8.70 | 16.15 | 4.60 |
| 49 | 11.47 | 7.48 | 12.56 | 14.48 | 12.45 | 22.32 | 6.06 |
| 50 | >30 | >30 | 6.89 | >30 | >30 | >30 | >30 |
| 51 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 52 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 53 | 3.47 | 3.41 | 2.13 | 3.39 | 3.41 | 3.54 | 2.73 |
| 54 | 3.52 | 3.52 | 2.48 | 3.38 | 4.49 | 4.54 | 3.40 |
| 55 | 5.48 | 4.05 | 4.94 | 5.43 | 6.27 | 5.95 | 4.03 |
| 56 | 4.02 | 3.76 | 4.06 | 3.88 | 4.38 | 4.02 | 2.46 |
| 57 | 2.89 | 4.04 | 2.59 | 2.35 | 1.48 | 0.80 | 3.01 |
| 58 | 2.49 | 2.21 | 1.98 | 2.53 | 3.01 | 2.70 | 1.55 |
| 59 | 2.40 | 2.43 | 1.58 | 2.43 | 2.27 | 2.51 | 1.75 |
Table 5.
Cytotoxicity (IC50 values in µM) of 60–66 in a panel of various cancer cell lines (NA = not active).
Table 5.
Cytotoxicity (IC50 values in µM) of 60–66 in a panel of various cancer cell lines (NA = not active).
| Cell Lines | 60 | 61 | 62 | 63 | 64 | 65 | 66 | GA–Me |
|---|
| SW1736 | NA | NA | NA. | 23.87 ± 1.3 | 11.18 ± 0.9 | 21.38 ± 1.9 | NA | 34.87 ± 1.2 |
| MCF-7 | NA | 16.7 ± 1.4 | 19.60 ± 1.4 | NA | 9.48 ± 1.4 | 20.11 ± 1.3 | NA | 22.14 ± 0.9 |
| LIPO | NA | NA | NA | 28.45 ± 2.1 | NA | 23.23 ± 1.3 | NA | 20.47 ± 1.1 |
| DLD-1 | NA | NA | NA | NA | NA | 23.18 ± 1.7 | NA | 26.12 ± 1.0 |
| A253 | NA | NA | NA | 27.25 ± 1.8 | 13.16 ± 0.9 | 19.70 ± 1.4 | NA | 19.42 ± 1.1 |
| 8505C | NA | NA | NA | NA | 21.97 ± 0.6 | 22.77 ± 1.4 | NA | 26.07 ± 1.3 |
| 518A2 | NA | NA | NA | 28.92 ± 2.0 | 25.95 ± 0.8 | 23.26 ± 1.2 | NA | 27.54 ± 1.0 |
| NiH3T3 | NA | NA | NA | NA | NA | 23.45 ± 0.1 | NA | 22.81 ± 0.6 |
Table 6.
Cytotoxicity (IC50 values in µM) of 68–74 in a panel of various cancer cell lines.
Table 6.
Cytotoxicity (IC50 values in µM) of 68–74 in a panel of various cancer cell lines.
| Cell Lines | GA | 68 | 69 | 70 | 71 | 72 | 73 | 74 |
|---|
| HepG2 | >50 | 18.18 | 13.41 | 26.03 | 36.52 | 15.67 | 7.90 | 2.90 |
| BEL-7402 | >50 | 7.85 | 9.22 | 6.03 | 8.20 | 19.92 | 7.37 | 2.94 |
Table 7.
Cytotoxicity (IC50 values in μM) of GA-1, GA-2 and GA-3 in A-549.
Table 7.
Cytotoxicity (IC50 values in μM) of GA-1, GA-2 and GA-3 in A-549.
| Cell Lines | GA-1 | GA-2 | GA-3 |
|---|
| A549 | 1.182 | 1.044 | 1.274 |
Table 8.
Cytotoxicity (IC50 values in μM) of 76–79 and β-CDODA-Me in a panel of various cancer cell lines.
Table 8.
Cytotoxicity (IC50 values in μM) of 76–79 and β-CDODA-Me in a panel of various cancer cell lines.
| Compound | 253JB-V | KU7 | Panc-1 | Panc-28 |
|---|
| 76 | 2.67 | 3.04 | 4.08 | 12.75 |
| 77 | 11.97 | 3.34 | 7.69 | 9.75 |
| 78 | 0.67 | 0.38 | 0.82 | 1.14 |
| 79 | 7.90 | 3.73 | 6.11 | 8.14 |
| β-CDODA-Me | 0.25 | 1.59 | 1.22 | 1.80 |
Table 9.
Cytotoxicity (IC50 values in μM) of 80–95, 97 in a panel of various cancer cell lines.
Table 9.
Cytotoxicity (IC50 values in μM) of 80–95, 97 in a panel of various cancer cell lines.
| Compound | 518A2 | 8505C | A2780 | A549 | DLD-1 | LIPO | MCF-7 | SW1736 |
|---|
| 80–85 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 86 | 18.33 | 19.28 | 28.83 | >30 | >30 | 28.74 | 21.87 | 16.56 |
| 87 | 29.82 | 27.69 | 14.84 | 26.62 | 29.56 | 24.80 | 28.68 | 27.00 |
| 88 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | 13.24 |
| 89 | >30 | 29.42 | >30 | >30 | >30 | >30 | >30 | 29.40 |
| 90–92 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 93 | >30 | >30 | 14.95 | >30 | >30 | >30 | >30 | 19.14 |
| 94, 95 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 97 | 23.69 | 24.30 | 10.39 | >30 | >30 | 25.52 | >30 | 16.98 |
Table 10.
Cytotoxicity (IC50 values in μM) of 98–112 in a panel of various cancer cell lines.
Table 10.
Cytotoxicity (IC50 values in μM) of 98–112 in a panel of various cancer cell lines.
| Cell Lines | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 |
|---|
| HepG-2 | 61.70 | >100 | 71.83 | 47.12 | >100 | >100 | >100 | 0.22 |
| | 106 | 107 | 108 | 109 | 110 | 111 | 112 | |
| HepG-2 | 59.98 | >100 | >100 | >100 | 88.68 | >100 | >100 | |
Table 11.
Cytotoxicity (IC50 values in μM) of 1, 12, 113–115 in a panel of various cancer cell lines.
Table 11.
Cytotoxicity (IC50 values in μM) of 1, 12, 113–115 in a panel of various cancer cell lines.
| Cell Lines | GA | 113 | 1 | 114 | 12 | 115 |
|---|
| 518A2 | 83.92 | 71.49 | 27.54 | 34.54 | 25.23 | 51.52 |
| 8505C | 86.50 | 78.52 | 26.07 | 33.88 | 24.58 | 52.80 |
| A2780 | 74.57 | 62.78 | 25.54 | 23.58 | 26.96 | 57.01 |
| A431 | 79.58 | 86.13 | 25.28 | 33.55 | 23.45 | 46.55 |
| A549 | 82.76 | 79.13 | 23.50 | 31.59 | 22.74 | 48.97 |
| DLD-1 | 81.21 | 90.50 | 26.12 | 31.73 | 28.14 | 52.80 |
| HCT-116 | 78.83 | 87.70 | 22.10 | 31.82 | 21.58 | 47.78 |
| HCT-8 | 78.85 | 88.76 | 24.36 | 31.34 | 43.42 | 44.32 |
| HT-29 | 80.09 | 90.30 | 27.54 | 23.89 | 22.14 | 44.32 |
| LIPO | 81.44 | 73.88 | 20.47 | 34.81 | 27.66 | 52.80 |
| MCF-7 | 84.70 | 90.19 | 22.14 | 34.37 | 18.61 | 48.97 |
| SW1736 | 76.93 | 72.47 | 34.87 | 32.35 | 13.37 | 45.48 |
| NIH3T3 | 18.52 | 68.70 | 22.81 | 42.22 | 23.66 | 43.16 |
Table 12.
Cytotoxicity (IC50 values in μM) of 116–130 in a panel of various cancer cell lines.
Table 12.
Cytotoxicity (IC50 values in μM) of 116–130 in a panel of various cancer cell lines.
| Compound | A549 | SKMEL | T98G | HS683 | U373 | PC3 | MCF7 | 816F10 |
|---|
| GA | >100 | 92 | 85 | 84 | 83 | 80 | 76 | 37 |
| 116 | 52 | >100 | 91 | 59 | 43 | 34 | 34 | 37 |
| 117 | 40 | >100 | >100 | 57 | 75 | 43 | 38 | 31 |
| 118 | 33 | 82 | 46 | 56 | 42 | 33 | 31 | 32 |
| 119 | 43 | 60 | 73 | 63 | 57 | 41 | 37 | 48 |
| 120 | 31 | >100 | >100 | 58 | 32 | 31 | 59 | 30 |
| 121 | 47 | 49 | 62 | 38 | 55 | 53 | 28 | 36 |
| 122 | 63 | 42 | 77 | 58 | 75 | 72 | 46 | 31 |
| 123 | 37 | 38 | 54 | 36 | 37 | 47 | 30 | 31 |
| 124 | 68 | 35 | 77 | 67 | 76 | 72 | 27 | 31 |
| 125 | 28 | 37 | 35 | 31 | 29 | 30 | 25 | 28 |
| 126 | 29 | 49 | 30 | 28 | 30 | 32 | 28 | 31 |
| 127 | 7 | 9 | 12 | 6 | 6 | 8 | 4 | 4 |
| 128 | 29 | 65 | 71 | 42 | 42 | 46 | 42 | 41 |
| 129 | 31 | 38 | 25 | 8 | 29 | 9 | 30 | 34 |
| 130 | 38 | 33 | 35 | 36 | 35 | 39 | 30 | 33 |
Table 13.
Cytotoxicity (GI50 values in μM) of 141–155 in a panel of various cancer cell lines.
Table 13.
Cytotoxicity (GI50 values in μM) of 141–155 in a panel of various cancer cell lines.
| Compound | A549 | DU145 | KB | Kbvin |
|---|
| GA | 61.2 ± 2.33 | 64.9 ± 0.505 | 61.2 ± 0.118 | 62.3 ± 1.41 |
| 141 | >70 | >70 | >70 | >70 |
| 142 | >70 | >70 | >70 | >70 |
| 143 | 19.4 ± 0.909 | 19.3 ± 0.292 | 14.6 ± 0.448 | 14.9 ± 0.471 |
| 144 | 34.2 ± 1.88 | 28.9 ± 0.921 | 17.5 ± 0.927 | 18.6 ± 0.931 |
| 145 | 23.3 ± 0.304 | 21.7 ± 0.402 | 16.9 ± 0.501 | 19.2 ± 0.497 |
| 146 | 44.0 ± 0.057 | 45.5 ± 0.666 | 39.9 ± 0.618 | 47.6 ± 1.06 |
| 147 | 18.3 ± 0.373 | 17.4 ± 0.619 | 15.3 ± 0.469 | 19.5 ± 1.33 |
| 148 | >70 | >70 | >70 | >70 |
| 149 | 19.6 ± 1.60 | 22.0 ± 0.546 | 16.0 ± 0.368 | 17.0 ± 0.377 |
| 150 | 15.0 ± 0.689 | 15.0 ± 0.363 | 14.2 ± 0.670 | 13.7 ± 1.25 |
| 151 | 46.7 ± 1.90 | 46.2 ± 0.697 | 45.5 ± 1.04 | 46.9 ± 0.230 |
| 152 | 46.1 ± 0.653 | 45.2 ± 1.27 | 41.3 ± 0.346 | 44.2 ± 0.280 |
| 153 | 19.0 ± 1.13 | 22.5 ± 0.606 | 17.8 ± 0.193 | 16.6 ± 0.591 |
| 154 | 34.5 ± 0.187 | 39.5 ± 1.05 | 30.7 ± 0.480 | 27.3 ± 0.338 |
| 155 | 41.5 ± 1.83 | 43.2 ± 1.61 | 38.4 ± 1.15 | 38.5 ± 0.956 |
Table 14.
Cytotoxicity (ED50 values in μM) of 156–166 in a panel of various cancer cell lines.
Table 14.
Cytotoxicity (ED50 values in μM) of 156–166 in a panel of various cancer cell lines.
| Compound | KB | KB-VIN | A549 | 1A9 | HCT-8 | ZR-751 | PC-3 | DU-145 | LN-Cap |
|---|
| GA | >21 | >21 | NA | >21 | 19.5 | NA | >21 | >21 | >21 |
| DZ | NA | NA | >52 | 33.9 | >52 | >52 | >52 | >52 | 51 |
| 156 | 1.6 | 2.5 | 2 | 0.9 | 1.7 | 2.8 | 1.4 | 3.1 | 0.6 |
| 157 | 0.8 | 2.8 | 2.2 | 0.8 | 1.9 | 3 | 1.1 | 3.6 | 2.8 |
| 158 | 0.9 | 1.9 | 2.8 | 1.6 | 2 | 1.9 | 2.8 | 9.9 | 6.5 |
| 159 | 6.2 | >15 | 15.5 | 5.9 | 2.6 | >15 | 7.4 | >15 | 1.9 |
| 160 | 1.8 | 1.7 | 1.7 | 1.1 | 2.7 | 5.2 | 3.3 | 5.8 | 1.1 |
| 161 | 2.9 | 13.2 | 3 | 1.8 | 4.9 | 8.8 | 3.5 | >15 | 6.8 |
| 162 | 3 | 8.7 | 3.2 | 1.3 | 2.2 | 2.7 | 1.6 | 2.7 | 4.4 |
| 163 | NA | NA | >14 | >14 | >14 | NA | >14 | >14 | >14 |
| 164 | 9.9 | NA | >14 | 13.3 | >14 | >14 | 14.1 | >14 | 14.1 |
| 165 | NA | NA | NA | >14 | >14 | NA | 14.1 | >14 | 14.1 |
| 166 | >14 | >14 | NA | NA | >14 | NA | >14 | 13 | >14 |
Table 15.
Cytotoxicity (IC50 values in μM) of 167–175 in a panel of various cancer cell lines.
Table 15.
Cytotoxicity (IC50 values in μM) of 167–175 in a panel of various cancer cell lines.
| Compound | 518A2 | 8505C | A253 | A549 | DLD-1 | Lipo | SW1736 |
|---|
| GA | 83.92 | 86.50 | 80.78 | 82.76 | 81.21 | 81.44 | 76.93 |
| 167 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 168 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 169 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
| 170 | 15.19 | 15.59 | 15.89 | 20.27 | 22.98 | 15.46 | 19.87 |
| 171 | 28.99 | >30 | >30 | >30 | >30 | >30 | 28.64 |
| 172 | 21.00 | 8.82 | 10.97 | 4.28 | 23.09 | 11.47 | 1.88 |
| 173 | 14.91 | 11.61 | 13.57 | 19.16 | 14.88 | 12.77 | 16.36 |
| 174 | 15.33 | 15.59 | 15.89 | 20.27 | 22.98 | 15.46 | 19.87 |
| 175 | >30 | >30 | >30 | >30 | >30 | >30 | >30 |
Table 16.
Cytotoxicity (IC50 values in μM) of 176–199 in a panel of various cancer cell lines.
Table 16.
Cytotoxicity (IC50 values in μM) of 176–199 in a panel of various cancer cell lines.
| Compound | MCF-7 | MDA-MB-231 | hTERT-RPE1 |
|---|
| GA | 75.66 ± 1.52 | 84.70 ± 1.73 | 63.41 ± 1.07 |
| 176 | 13.64 ± 0.93 | 5.03 ± 0.82 | 17.32 ± 1.21 |
| 177 | 22.46 ± 1.26 | 8.14 ± 0.76 | 22.80 ± 0.97 |
| 178 | 20.29 ± 1.47 | 14.38 ± 0.52 | 29.63 ± 1.16 |
| 179 | 24.45 ± 1.36 | 14.46 ± 0.58 | 28.41 ± 0.87 |
| 180 | 8.54 ± 0.67 | 7.31 ± 0.16 | 18.59 ± 0.54 |
| 181 | 19.27 ± 1.01 | 9.41 ± 1.03 | 21.11 ± 0.73 |
| 182 | 14.90 ± 0.75 | 20.84 ± 1.20 | 24.09 ± 0.88 |
| 183 | 19.30 ± 0.98 | 23.15 ± 1.07 | 22.88 ± 0.68 |
| 192 | 6.00 ± 0.43 | 3.52 ± 0.61 | 10.36 ± 0.80 |
| 193 | 1.88 ± 0.20 | 1.37 ± 0.18 | 4.93 ± 0.36 |
| 194 | 8.62 ± 0.23 | 5.36 ± 0.44 | 16.28 ± 0.51 |
| 195 | 8.45 ± 0.32 | 3.49 ± 0.61 | 12.33 ± 0.46 |
| 196 | 7.24 ± 0.30 | 6.43 ± 0.84 | 8.48 ± 0.73 |
| 197 | 6.02 ± 0.35 | 6.27 ± 0.24 | 6.33 ± 0.19 |
| 198 | 2.65 ± 0.12 | 2.31 ± 0.65 | 5.65 ± 1.02 |
| 199 | 2.42 ± 0.23 | 1.86 ± 0.29 | 7.08 ± 0.73 |
Table 17.
Cytotoxicity (IC50 values in μM) of 200–206, 209 and 210 in a panel of various cancer cell lines.
Table 17.
Cytotoxicity (IC50 values in μM) of 200–206, 209 and 210 in a panel of various cancer cell lines.
| Compound | HepG2 | DU-145 | MDA-MB-231 |
|---|
| GA | 74.35 ± 2.03 | 69.40 ± 2.37 | 72.65 ± 1.67 |
| 200 | >100 | 21.59 ± 3.22 | 24.66 ± 2.71 |
| 201 | >100 | >100 | 89.40 ± 2.85 |
| 202 | >100 | >100 | >100 |
| 203 | 10.01 ± 2.29 | 11.96 ± 1.42 | 17.80 ± 1.76 |
| 204 | >100 | >100 | 79.3 ± 2.34 |
| 205 | 36.37 ± 1.89 | >100 | 40.65 ± 2.11 |
| 206 | >100 | >100 | >100 |
| 209 | >100 | >100 | >100 |
| 210 | >100 | >100 | >100 |















 , 1N NaOH (for R2 = Me), 5N KOH (for R2 = Ph); (b) GA, EDCI, DMAP, CH2Cl2.
, 1N NaOH (for R2 = Me), 5N KOH (for R2 = Ph); (b) GA, EDCI, DMAP, CH2Cl2.
 , 1N NaOH (for R2 = Me), 5N KOH (for R2 = Ph); (b) GA, EDCI, DMAP, CH2Cl2.
, 1N NaOH (for R2 = Me), 5N KOH (for R2 = Ph); (b) GA, EDCI, DMAP, CH2Cl2.