Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis
Abstract
:1. Introduction
2. Results
2.1. Synthesis of PNAs 1–4
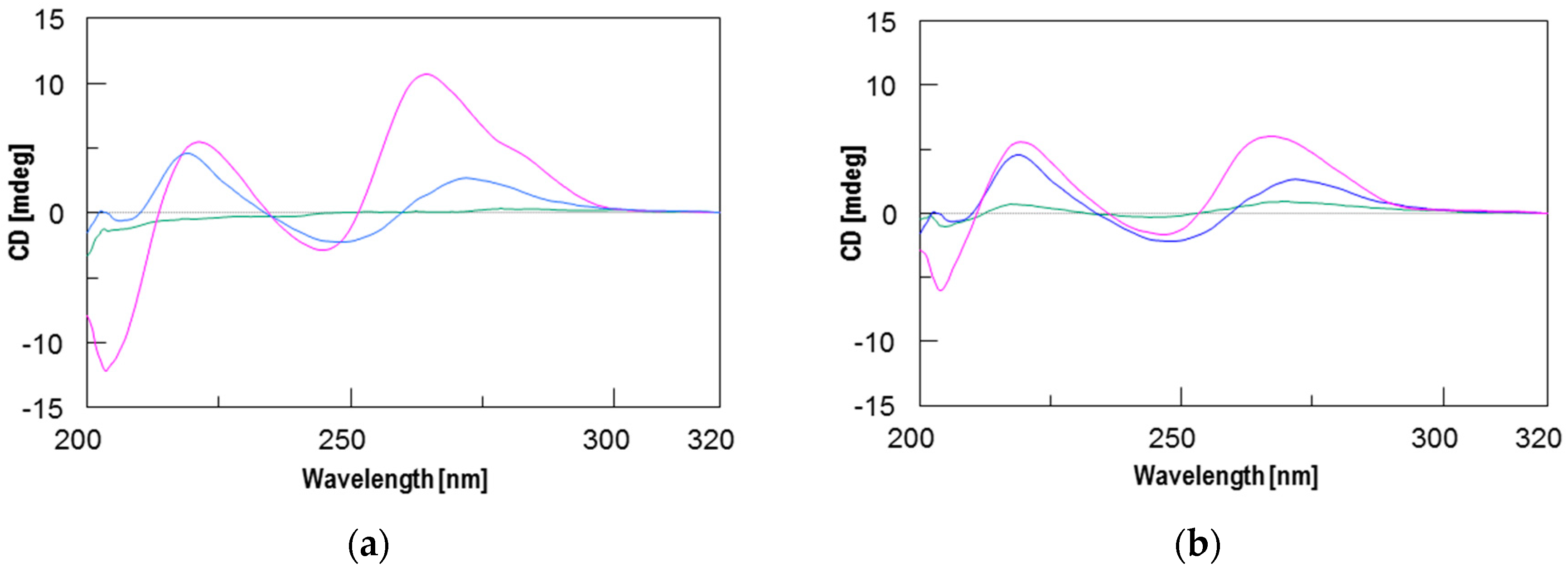
2.2. Circular Dichroism (CD) and CD Melting Analyses
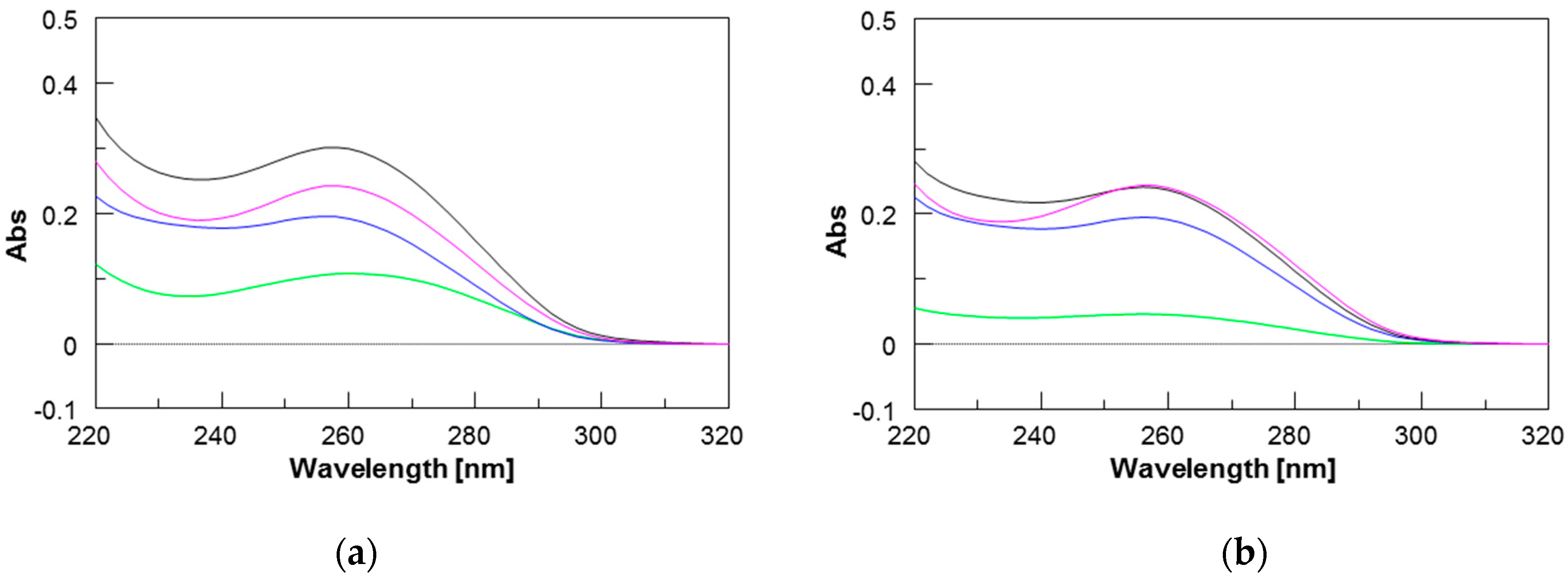
2.3. UV Studies
2.4. Molecular Modelling Studies
2.5. Biological Activity
3. Discussion
4. Materials and Methods
4.1. General Methods
4.2. DNA Synthesis and Analysis
4.3. PNA Synthesis and Analysis
4.4. Preparation of DNA/PNA Heteroduplexes (Annealing Procedure)
4.5. UV
4.6. CD and CD Melting Studies
4.7. Molecular Dynamics (MD) Simulations
4.8. Cell Line, Construct, and Transfections
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904. [Google Scholar] [CrossRef]
- Heda, G.D.; Marino, C.R. Surface expression of the cystic fibrosis transmembrane conductance regulator mutant deltaf508 is markedly upregulated by combination treatment with sodium butyrate and low temperature. Biochem. Biophys. Res. Commun. 2000, 271, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Amato, F.; Seia, M.; Giordano, S.; Elce, A.; Zarrilli, F.; Castaldo, G.; Tomaiuolo, R. Gene mutation in microRNA target sites of CFTR gene: A novel pathogenetic mechanism in cystic fibrosis? PLoS ONE 2013, 8, e60448. [Google Scholar] [CrossRef] [PubMed]
- Gillen, A.E.; Gosalia, N.; Leir, S.H.; Harris, A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 2011, 438, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Karp, P.H.; Osterhaus, S.R.; Jiang, P.; Wohlford-Lenane, C.; Lennox, K.A.; Jacobi, A.M.; Praekh, K.; Rose, S.D.; Behlke, M.A.; et al. Post-transcriptional Regulation of CFTR Expression and Function by MicroRNAs. Am. J. Respir. Cell. Mol. Biol. 2013, 49, 544–551. [Google Scholar] [CrossRef]
- Oglesby, I.K.; Chotirmall, S.H.; McElvaney, N.G.; Greene, C.M. Regulation of Cystic Fibrosis Transmembrane Conductance Regulator by MicroRNA-145, -223, and -494 Is Altered in ΔF508 Cystic Fibrosis Airway Epithelium. J. Immunol. 2013, 190, 3354–3362. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.; Nuovo, G.J.; Crawford, M.; Boyaka, P.N.; Kirkby, S.; Nana-Sinkam, S.P.; Cormet-Boyaka, E. MiR-101 and miR-144 Regulate the Expression of the CFTR Chloride Channel in the Lung. PLoS ONE 2012, 7, e50837. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Karp, P.H.; Jiang, P.; Ostedgaard, L.S.; Walz, A.E.; Fisher, J.T.; Keshavjee, S.; Lennox, K.A.; Jacobi, A.M.; Rose, S.D.; et al. A microRNA network regulates expression and biosynthesis of wild-type and F508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. USA 2012, 109, 13362–13367. [Google Scholar] [CrossRef] [PubMed]
- Megiorni, F.; Cialfi, S.; Dominici, C.; Quattrucci, S.; Pizzuti, A. Synergistic Post-Transcriptional Regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 Specific Binding. PLoS ONE 2011, 6, e26601. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. Mirbase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Amato, F.; Tomaiuolo, R.; Borbone, N.; Elce, A.; Amato, J.; D’Errico, S.; De Rosa, G.; Mayol, L.; Piccialli, G.; Oliviero, G.; et al. Design, synthesis and biochemical investigation, by in vitro luciferase reporter system, of peptide nucleic acids as new inhibitors of miR-509–3p involved in the regulation of cystic fibrosis disease-gene expression. Med. Chem. Commun. 2014, 5, 68–71. [Google Scholar] [CrossRef]
- Amato, F.; Tomaiuolo, R.; Nici, F.; Borbone, N.; Elce, A.; Catalanotti, B.; D’Errico, S.; Morgillo, C.M.; De Rosa, G.; Mayol, L.; et al. Exploitation of a very small peptide nucleic acid as a new inhibitor of miR-509–3p involved in the regulation of cystic fibrosis disease-gene expression. Biomed. Res. Int. 2014, 2014, 610718. [Google Scholar] [CrossRef] [PubMed]
- Amato, J.; Oliviero, G.; De Pauw, E.; Gabelica, V. Hybridization of short complementary PNAs to G-quadruplex forming oligonucleotides: An electrospray mass spectrometry study. Biopolymers 2009, 91, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Amato, J.; Pagano, B.; Borbone, N.; Oliviero, G.; Gabelica, V.; Pauw, E.D.; D'Errico, S.; Piccialli, V.; Varra, M.; Giancola, C.; et al. Targeting G-quadruplex structure in the human c-kit promoter with short PNA sequences. Bioconj. Chem. 2011, 22, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Amato, J.; Stellato, M.I.; Pizzo, E.; Petraccone, L.; Oliviero, G.; Borbone, N.; Piccialli, G.; Orecchia, A.; Bellei, B.; Castiglia, D.; et al. PNA as a potential modulator of col7a1 gene expression in dominant dystrophic epidermolysis bullosa: A physico-chemical study. Mol. Biosyst. 2013, 9, 3166–3174. [Google Scholar] [CrossRef] [PubMed]
- Avitabile, C.; Saviano, M.; D'Andrea, L.; Bianchi, N.; Fabbri, E.; Brognara, E.; Gambari, R.; Romanelli, A. Targeting pre-miRNA by peptide nucleic acids: A new strategy to interfere in the miRNA maturation. Artif. DNA: PNA&XNA 2012, 3, 88–96. [Google Scholar]
- Brognara, E.; Fabbri, E.; Montagner, G.; Gasparello, J.; Manicardi, A.; Corradini, R.; Bianchi, N.; Finotti, A.; Breveglieri, G.; Borgatti, M.; et al. High levels of apoptosis are induced in human glioma cell lines by co-administration of peptide nucleic acids targeting miR-221 and miR-222. Int. J. Oncol. 2016, 48, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Hyrup, B.; Nielsen, P.E. Peptide nucleic acids (PNA): Synthesis, properties and potential applications. Bioorg. Med. Chem. 1996, 4, 5–23. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Musumeci, D.; De Cristofaro, A.; Capasso, D.; Di Gaetano, S.; Bucci, E.M.; Pedone, C. Alternate dab-aegPNAs: Synthesis, nucleic acid binding studies and biological activity. Mol. Biosyst. 2010, 6, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Ricci, A.; Bucci, E.M.; Pedone, C. Synthesis, biological evaluation and supramolecular assembly of novel analogues of peptidyl nucleosides. Mol. Biosyst. 2011, 7, 1773–1778. [Google Scholar] [CrossRef] [PubMed]
- Pinto, B.; Rusciano, G.; D’Errico, S.; Borbone, N.; Sasso, A.; Piccialli, V.; Mayol, L.; Oliviero, G.; Piccialli, G. Synthesis and label free characterization of a bimolecular PNA homo quadruplex. BBA-Gen. Subj. 2016, 1861, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Viart, V.; Bergougnoux, A.; Bonini, J.; Varilh, J.; Chiron, R.; Tabary, O.; Molinari, N.; Claustres, M.; Taulan-Cadars, M. Transcription factors and miRNAs that regulate fetal to adult CFTR expression change are new targets for cystic fibrosis. Eur. Respir. J. 2014, 45, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.K.; Orum, H.; Nielsen, P.E.; Norden, B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry 1997, 36, 5072–5077. [Google Scholar] [CrossRef] [PubMed]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the watson-crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.C.; Thomson, S.A.; Veal, J.M.; Davis, D.G. NMR solution structure of a peptide nucleic acid complexed with RNA. Science 1994, 265, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, J.T.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; et al. Amber 14; University of California: San Francisco, CA, USA, 2015. [Google Scholar]
- Lavery, R.; Moakher, M.; Maddocks, J.H.; Petkeviciute, D.; Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 2009, 37, 5917–5929. [Google Scholar] [CrossRef] [PubMed]
- Kiliszek, A.; Banaszak, K.; Dauter, Z.; Rypniewski, W. The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs. Nucleic Acids Res. 2016, 44, 1937–1943. [Google Scholar] [CrossRef] [PubMed]
- Soliva, R.; Sherer, E.; Luque, F.J.; Laughton, C.A.; Orozco, M. Molecular dynamics simulations of PNA·DNA and PNA·RNA duplexes in aqueous solution. J. Am. Chem. Soc. 2000, 122, 5997–6008. [Google Scholar] [CrossRef]
- Tanaka, Y.; Fujii, S.; Hiroaki, H.; Sakata, T.; Tanaka, T.; Uesugi, S.; Tomita, K.; Kyogoku, Y. A’-form RNA double helix in the single crystal structure of r (UGAGCUUCGGCUC). Nucleic Acids Res. 1999, 27, 949–955. [Google Scholar] [CrossRef]
- Autiero, I.; Saviano, M.; Langella, E. Molecular dynamics simulations of PNA-PNA and PNA-DNA duplexes by the use of new parameters implemented in the GROMACS package: A conformational and dynamics study. Phys. Chem. Chem. Phys. 2014, 16, 1868–1874. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Hatcher, E.; Balaeff, A.; Beratan, D.N.; Gil, R.R.; Madrid, M.; Achim, C. Solution structure of a peptide nucleic acid duplex from NMR data: Features and limitations. J. Am. Chem. Soc. 2008, 130, 13264–13273. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.M.; Wampole, M.E.; Chen, C.P.; Sethi, D.; Singh, A.; Dupradeau, F.Y.; Wang, F.; Gray, B.D.; Thakur, M.L.; Wickstrom, E.; et al. Effects of hypoxanthine substitution in peptide nucleic acids targeting kras2 oncogenic mRNA molecules: Theory and experiment. J. Phys. Chem. B 2013, 117, 11584–11595. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Nilsson, L. Molecular dynamics of duplex systems involving PNA: Structural and dynamical consequences of the nucleic acid backbone. J. Am. Chem. Soc. 1998, 120, 619–631. [Google Scholar] [CrossRef]
- Greene, C.M.; Hartl, D. Developmental control of CFTR: From bioinformatics to novel therapeutic approaches. Eur. Respir. J. 2015, 45, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Hasuwa, H.; Ueda, J.; Ikawa, M.; Okabe, M. MiR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 2013, 341, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14sb: Improving the accuracy of protein side chain and backbone parameters from ff99sb. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.; Marchan, I.; Svozil, D.; Sponer, J.; Cheatham, T.E., 3rd; Laughton, C.A.; Orozco, M. Refinement of the amber force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [PubMed]
- Zgarbova, M.; Otyepka, M.; Sponer, J.; Mladek, A.; Banas, P.; Cheatham, T.E., 3rd; Jurecka, P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef] [PubMed]
- Craft, J.W., Jr.; Legge, G.B. An AMBER/DYANA/MOLMOL phosphorylated amino acid library set and incorporation into NMR structure calculations. J. Biomol. NMR 2005, 33, 15–24. [Google Scholar] [CrossRef] [PubMed]
- The PyMol Molecular Graphics System; Version 1.8; Schrödinger, LLC: New York, NY, USA.
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 27–38. [Google Scholar] [CrossRef]
Sample Availability: Samples of the DNA and PNA molecules reported in this study are available from the authors. |




| Name | Sequence 1,2 |
|---|---|
| 3’UTR of CFTR mRNA (RNA) | G-A-A-G-A-A-G-C-A-C-C-A-A-U-C-A-U-G-A |
| DNA model sequence (ODN) | G-A-A-G-C-A-C-C-A-A-T-C-A |
| PNA 1 (C → N) | G-S(P)-S(P)-G–c-t-t-c-g-t-g-g-t-t-a-g-t |
| PNA 2 (C → N) | G-S(P)-S(P)-G–g-g-t-t-a-g-t |
| PNA 3 (C → N) | G-S(P)-S(P)-G–c-a-g-t-t-g-t-c-t-g-t-g-t |
| PNA 4 (C → N) | G-S(P)-S(P)-G–t-t-g-g-a-g-t |
| Name | Shift | Slide | Rise | Tilt | Roll | Twist |
|---|---|---|---|---|---|---|
| 1/RNA | −0.7 (0.2) | −1.9 (0.2) | 3.3 (0.2) | 1.0 (1.4) | 2.4 (1.9) | 23.6 (1.7) |
| 2/RNA | −0.9 (0.3) | −1.7 (0.2) | 3.3 (0.1) | 1.3 (1.5) | 3.9 (1.9) | 22.6 (1.3) |
| PNA/RNA (NMR) 1 | 0.3 (0.3) | −1.4 (0.6) | 3.2 (0.4) | −2.8 (1.3) | 4.6 (4.3) | 29.4 (3.8) |
| PNA/RNA (MD) 2 | - | - | - | - | - | 24 |
| PNA/RNA (X-RAY) 3 | −0.8 (0.4) | −2.1 (0.3) | 3.3 (0.1) | 0.1 (1.3) | 6.9 (3.5) | 25.0 (1.6) |
| A-RNA 4 | - | - | 2.8 | - | - | 32.7 |
| Torsional PNA Angles | |||||||||
| Name | α | β | γ | δ | ε | ω | χ1 | χ2 | χ3 |
| 1/RNA | –120 (46.0) | 78 (10.6) | 71 (2.5) | 95 (1.8) | –177 (42.9) | –146 (10.0) | –2 (1.2) | –164 (2.2) | 84 (2.8) |
| 2/RNA | –136 (43.2) | 76 (4.1) | 79 (3.9) | 96 (2.0) | –162 (56.6) | –172 (10.6) | –3 (1.6) | –152 (18.0) | 77 (12.6) |
| PNA/RNA (NMR) 1 | 160 (10.1) | 68 (2.4) | 81 (5.0) | 59 (16.1) | –104 (6.7) | –177 (3.2) | 12 (3.3) | –118 (8.6) | 49 (10.2) |
| PNA/RNA (XRAY) 2 | –177 (83.5) | 68 (8.8) | 71 (7.5) | 95 (5.2) | –123 (81.0) | –178 (10.5) | 5 (5.1) | –173 (4.0) | 82 (6.6) |
| Torsional RNA Angles | |||||||||
| Name | α | β | γ | δ | ε | ζ | χ | ||
| 1/RNA | –77 (1.5) | 172 (1.7) | 68 (1.5) | 79 (0.9) | –162 (2.7) | –72 (2.9) | –160 (3.1) | ||
| 2/RNA | –78 (1.6) | 173 (1.9) | 67 (1.6) | 79 (1.3) | –164 (2.7) | –73 (2.4) | –156 (2.8) | ||
| PNA/RNA (NMR) 1 | –68 (6.7) | 171 (13.8) | 58 (1.2) | 79 (3.6) | –149 (23.8) | –73 (11.6) | –168 (4.4) | ||
| PNA/RNA (X-RAY) 2 | –80 (4.9) | 177 (5.4) | 63 (4.9) | 79 (6.2) | –158 (5.5) | –71 (6.6) | –164 (4.9) | ||
| A-RNA 3 | –79 (43.5) | 172 (19.9) | 64 (34.7) | 80 (9.53) | –152 (17.6) | –76 (24.7) | –162 (9.76) | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarrilli, F.; Amato, F.; Morgillo, C.M.; Pinto, B.; Santarpia, G.; Borbone, N.; D’Errico, S.; Catalanotti, B.; Piccialli, G.; Castaldo, G.; et al. Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis. Molecules 2017, 22, 1144. https://doi.org/10.3390/molecules22071144
Zarrilli F, Amato F, Morgillo CM, Pinto B, Santarpia G, Borbone N, D’Errico S, Catalanotti B, Piccialli G, Castaldo G, et al. Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis. Molecules. 2017; 22(7):1144. https://doi.org/10.3390/molecules22071144
Chicago/Turabian StyleZarrilli, Federica, Felice Amato, Carmine Marco Morgillo, Brunella Pinto, Giuliano Santarpia, Nicola Borbone, Stefano D’Errico, Bruno Catalanotti, Gennaro Piccialli, Giuseppe Castaldo, and et al. 2017. "Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis" Molecules 22, no. 7: 1144. https://doi.org/10.3390/molecules22071144
APA StyleZarrilli, F., Amato, F., Morgillo, C. M., Pinto, B., Santarpia, G., Borbone, N., D’Errico, S., Catalanotti, B., Piccialli, G., Castaldo, G., & Oliviero, G. (2017). Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis. Molecules, 22(7), 1144. https://doi.org/10.3390/molecules22071144









