Biosynthesis of S-Adenosylmethionine by Magnetically Immobilized Escherichia coli Cells Highly Expressing a Methionine Adenosyltransferase Variant
Abstract
:1. Introduction
2. Results and Discussion
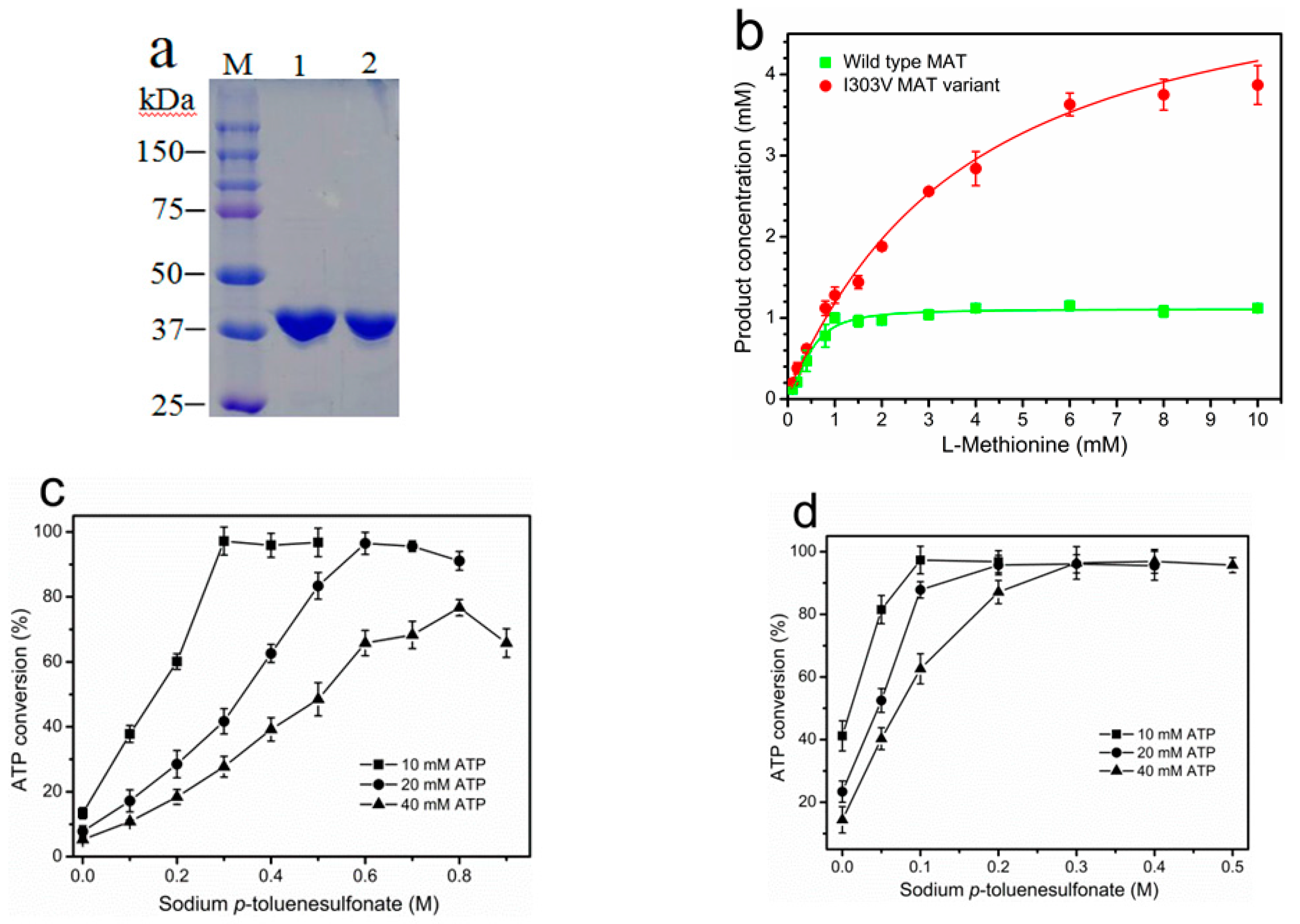
2.1. Purification and Properties of Recombinant Wild-Type and I303V MAT
2.2. Product Inhibition of Wild-Type and I303V MAT
2.3. Fermentation of Recombinant E. coli Expressing I303V MAT
2.4. Permeabilization of Free Whole-Cells of E. coli
2.5. Preparation of Magnetically Immobilized Cells
2.6. Thermal Inactivation of Free and Immobilized Cells
2.7. Reusability of Magnetically Immobilized Cells for SAM Production
3. Materials and Methods
3.1. Materials
3.2. Preparation of Recombinant, Wild-Type, and I303V MAT
3.3. Permeabilization of E. coli cells
3.4. Preparation of Nonmagnetically and Magnetically Immobilized Cells
3.5. Spectrophotometric Assay for MAT Activity
3.6. High-Performance Liquid Chromatography (HPLC) Analysis
3.7. Thermal Stability of the Free and Magnetically Immobilized Cells
3.8. Reusability of the Magnetically Immobilized Cells for SAM Biosynthesis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Grillo, M.A.; Colombatto, S. S-adenosylmethionine and its products. Amino Acids 2008, 34, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Fontecave, M.; Atta, M.; Mulliez, E. S-adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 2004, 29, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Noureddin, M.; Mato, J.M.; Lu, S.C. Nonalcoholic fatty liver disease: Update on pathogenesis, diagnosis, treatment and the role of S-adenosylmethionine. Exp. Biol. Med. 2015, 240, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Hosea Blewett, H.J. Exploring the mechanisms behind S-adenosylmethionine (SAMe) in the treatment of osteoarthritis. Crit. Rev. Food Sci. Nutr. 2008, 48, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Shippy, R.A.; Mendez, D.; Jones, K.; Cergnul, I.; Karpiak, S.E. S-adenosylmethionine (SAM-e) for the treatment of depression in people living with HIV/AIDS. BMC Psychiatry 2004, 4, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Tai, J.; Roessner, C.A.; Scott, A.I. Enzymatic synthesis of S-adenosyl-l-methionine on the preparative scale. Bioorg. Med. Chem. 1996, 4, 2179–2185. [Google Scholar] [CrossRef]
- Luo, Y.; Yuan, Z.; Luo, G.; Zhao, F. Expression of secreted His-tagged S-adenosylmethionine synthetase in the methylotrophic yeast Pichia pastoris and its characterization, one-step purification, and immobilization. Biotechnol. Prog. 2008, 24, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Qian, J.; Zhuang, Y.; Zhang, S.; Li, Y. Progress in the research of S-adenosyl-l-methionine production. Appl. Microbiol. Biotechnol. 2013, 97, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, Z.; Cai, H.; Zhou, C. Progress in the microbial production of S-adenosyl-l-methionine. World J. Microbiol. Biotechnol. 2016, 32, 153. [Google Scholar] [CrossRef] [PubMed]
- Kamarthapu, V.; Ragampeta, S.; Rao, K.V.; Reddy, V.D. Engineered Pichia pastoris for enhanced production of S-adenosylmethionine. AMB Express 2013, 3, 40. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Shi, F.; Hang, B.; Huang, L.; Cai, J.; Xu, Z. The improvement of SAM accumulation by integrating the endogenous methionine adenosyltransferase gene SAM2 in genome of the industrial Saccharomyces cerevisiae strain. Appl. Biochem. Biotechnol. 2016, 178, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Mizunuma, M.; Fujii, T.; Iefuji, H. A genetic method to enhance the accumulation of S-adenosylmethionine in yeast. Appl. Microbiol. Biotechnol. 2017, 101, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Y.; Wang, Z.; Dou, J.; Wang, H.; Zhou, C. Elevated intracellular acetyl-CoA availability by ACS2 overexpression and MLS1 deletion combined with metK1 introduction enhanced SAM accumulation in Saccharomyces cerevisiae. Biochem. Eng. J. 2016, 107, 26–34. [Google Scholar] [CrossRef]
- Hu, X.; Chu, J.; Zhang, S.; Zhuang, Y.; Wang, Y.; Zhu, S.; Zhu, Z.; Yuan, Z. A novel feeding strategy during the production phase for enhancing the enzymatic synthesis of S-adenosyl-l-methionine by methylotrophic Pichia pastoris. Enzym. Microb. Technol. 2007, 40, 669–674. [Google Scholar] [CrossRef]
- He, J.; Deng, J.; Zheng, Y.; Gu, J. A synergistic effect on the production of S-adenosyl-l-methionine in Pichia pastoris by knocking in of S-adenosyl-l-methionine synthase and knocking out of cystathionine-beta synthase. J. Biotechnol. 2006, 126, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Qian, J.; Chu, J.; Wang, Y.; Zhuang, Y.; Zhang, S. Optimization of l-methionine feeding strategy for improving S-adenosyl-l-methionine production by methionine adenosyltransferase overexpressed Pichia pastoris. Appl. Microbiol. Biotechnol. 2009, 83, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Su, E.; Fang, G.; Ren, Y.; Wei, D. A new fermentation strategy for S-adenosylmethionine production in recombinant Pichia pastoris. Biochem. Eng. J. 2008, 41, 74–78. [Google Scholar] [CrossRef]
- Zhao, W.; Hang, B.; Zhu, X.; Wang, R.; Shen, M.; Huang, L.; Xu, Z. Improving the productivity of S-adenosyl-l-methionine by metabolic engineering in an industrial Saccharomyces cerevisiae strain. J. Biotechnol. 2016, 236, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Kisukuri, C.M.; Andrade, L.H. Production of chiral compounds using immobilized cells as a source of biocatalysts. Org. Biomol. Chem. 2015, 13, 10086–10107. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Tai, J.; Roessner, C.A.; Scott, A.I. Overcoming product inhibition of S-adenosyl-l-methionine (SAM) synthetase: Preparation of SAM on the 30 mM scale. Bioorg. Med. Chem. Lett. 1995, 5, 2203–2206. [Google Scholar] [CrossRef]
- Shi, S.; Qu, Y.; Tan, L.; Ma, F. Biosynthesis of 1,2-dihydroxydibenzofuran by magnetically immobilized cells of Escherichia coli expressing phenol hydroxylase in liquid-liquid biphasic systems. Bioresour. Technol. 2015, 197, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhang, W.; Yang, Z.; Yang, X.; Wang, N.; Yu, X. Novel magnetic cross-linked cellulase aggregates with a potential application in lignocellulosic biomass bioconversion. Molecules 2017, 22, 269. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.L.; Chaudhary, R.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Immobilization of β-glucosidase on a magnetic nanoparticle improves thermostability: Application in cellobiose hydrolysis. Bioresour. Technol. 2013, 135, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Qu, Y.; Ma, F.; Zhou, J. Bioremediation of coking wastewater containing carbazole, dibenzofuran and dibenzothiophene by immobilized naphthalene-cultivated Arthrobacter sp. W1 in magnetic gellan gum. Bioresour. Technol. 2014, 166, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, R.M.; Andrade Neto, D.M.; Galvão, W.S.; Rios, N.S.; Carvalho, A.C.L.; Correa, M.A.; Bohn, F.; Fernandez-Lafuente, R.; Fechine, P.B.A.; Mattos, M.C.; et al. Design of a lipase-nano particle biocatalysts and its use in the kinetic resolution of medicament precursors. Biochem. Eng. J. 2017, 125, 104–115. [Google Scholar] [CrossRef]
- Costa, V.M.; de Souza, M.C.M.; Fechine, P.B.A.; Macedo, A.C.; Gonҫalves, L.R.B. Nanobiocatalytic systems based on lipase-Fe3O4 and conventional systems for isoniazid synthesis: A comparative study. Braz. J. Chem. Eng. 2016, 33, 661–673. [Google Scholar] [CrossRef]
- Kamarthapu, V.; Rao, K.V.; Srinivas, P.N.; Reddy, G.B.; Reddy, V.D. Structural and kinetic properties of Bacillus subtilis S-adenosylmethionine synthetase expressed in Escherichia coli. Biochim. Biophys. Acta 2008, 1784, 1949–1958. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chu, J.; Wang, Y.H.; Zhang, S.L.; Zhuang, Y.P.; Yuan, Z.Y. Purification and properties of Saccharomyces cerevisiae S-adenosylmethionine synthetase expressed in recombinant Pichia pastoris. World J. Microbiol. Biotechnol. 2008, 24, 789–796. [Google Scholar] [CrossRef]
- Sakata, S.F.; Shelly, L.L.; Ruppert, S.; Schutz, G.; Chou, J.Y. Cloning and expression of murine S-adenosylmethionine synthetase. J. Biol. Chem. 1993, 268, 13978–13986. [Google Scholar] [PubMed]
- Markham, G.D.; Hafner, E.W.; Tabor, C.W.; Tabor, H. S-adenosylmethionine synthetase from Escherichia coli. J. Biol. Chem. 1980, 255, 9082–9092. [Google Scholar] [PubMed]
- Dippe, M.; Brandt, W.; Rost, H.; Schmidt, J.; Wessjohann, L.A. Rationally engineered variants of S-adenosylmethionine (SAM) synthase: Reduced product inhibition and synthesis of artificial cofactor homologues. Chem. Commun. 2015, 51, 3637–3640. [Google Scholar] [CrossRef] [PubMed]
- Komoto, J.; Yamada, T.; Takata, Y.; Markham, G.D.; Takusaqawa, F. Crystal structure of the S-adenosylmethionine synthetase ternary complex: A novel catalytic mechanism of S-adenosylmethionine synthesis from ATP and Met. Biochemistry 2004, 43, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Avinash, VS.; Chauhan, P.D.; Gaikwad, S.; Pundle, A. Biotransformation of penicillin V to 6-aminopenicillanic acid using immobilized whole cells of E. coli expressing a highly active penicillin V acylase. Prep. Biochem. Biotechnol. 2017, 47, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Wei, D.; Song, Q.; Zhao, X. Immobilization of whole cell permeabilized penicillin G acylase from alcaligenes faecalis using pore matrix crosslinked with glutaraldehyde. Biotechnol. Lett. 2006, 28, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Prabhune, A.A.; Rao, B.S.; Pundle, A.V.; Sivaraman, H. Immobilization of permeabilized Escherichia coli cells with penicillin acylase activity. Enzym. Microb. Technol. 1992, 14, 161–163. [Google Scholar] [CrossRef]
- Norouzian, D.; Javadpour, S.; Moazami, N.; Akbarzadeh, A. Immobilization of whole cell penicillin G acylase in open pore gelatin matrix. Enzym. Microb. Technol. 2002, 30, 26–29. [Google Scholar] [CrossRef]
- Wang, X.; Gai, Z.; Yu, B.; Feng, J.; Xu, C.; Yuan, Y.; Lin, Z.; Xu, P. Degradation of carbazole by microbial cells immobilized in magnetic gellan gum gel beads. Appl. Environ. Microbiol. 2007, 73, 6421–6428. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lopez, L.; Pedrero, S.G.; Lopez-Carrobles, N.; Gorines, B.C.; Virgen-ortíz, J.J.; Fernandez-Lafuente, R. Effect of protein load on stability of immobilized enzymes. Enzym. Microb. Technol. 2017, 98, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, Z.; Milosavic, N.; Bezbradica, D.; Jakovljevic, Z.; Prodanovic, R. Immobilization of lipase from Candida rugosa on Eupergit® C supports by covalent attachment. Biochem. Eng. J. 2006, 30, 269–278. [Google Scholar] [CrossRef]
Sample Availability: Not available. |





| Cell Loading (g/L Support) | Activity of Immobilized Cells (U/L Support) | Activity Recovery (%) |
|---|---|---|
| 10 | 1529.3 | 86.4 |
| 20 | 2906.3 | 82.1 |
| 30 | 4152.4 | 78.2 |
| 40 | 3957.7 | 55.9 |
| 50 | 4053.5 | 45.8 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Zheng, T.; Chang, X. Biosynthesis of S-Adenosylmethionine by Magnetically Immobilized Escherichia coli Cells Highly Expressing a Methionine Adenosyltransferase Variant. Molecules 2017, 22, 1365. https://doi.org/10.3390/molecules22081365
Yin C, Zheng T, Chang X. Biosynthesis of S-Adenosylmethionine by Magnetically Immobilized Escherichia coli Cells Highly Expressing a Methionine Adenosyltransferase Variant. Molecules. 2017; 22(8):1365. https://doi.org/10.3390/molecules22081365
Chicago/Turabian StyleYin, Chunli, Tao Zheng, and Xin Chang. 2017. "Biosynthesis of S-Adenosylmethionine by Magnetically Immobilized Escherichia coli Cells Highly Expressing a Methionine Adenosyltransferase Variant" Molecules 22, no. 8: 1365. https://doi.org/10.3390/molecules22081365
APA StyleYin, C., Zheng, T., & Chang, X. (2017). Biosynthesis of S-Adenosylmethionine by Magnetically Immobilized Escherichia coli Cells Highly Expressing a Methionine Adenosyltransferase Variant. Molecules, 22(8), 1365. https://doi.org/10.3390/molecules22081365




