The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations
Abstract
:1. Glycosylations Form a Huge Family of Co- and Post-Translational Modifications
2. O-GlcNAcylation Differs from Other Glycosylations in Many Ways
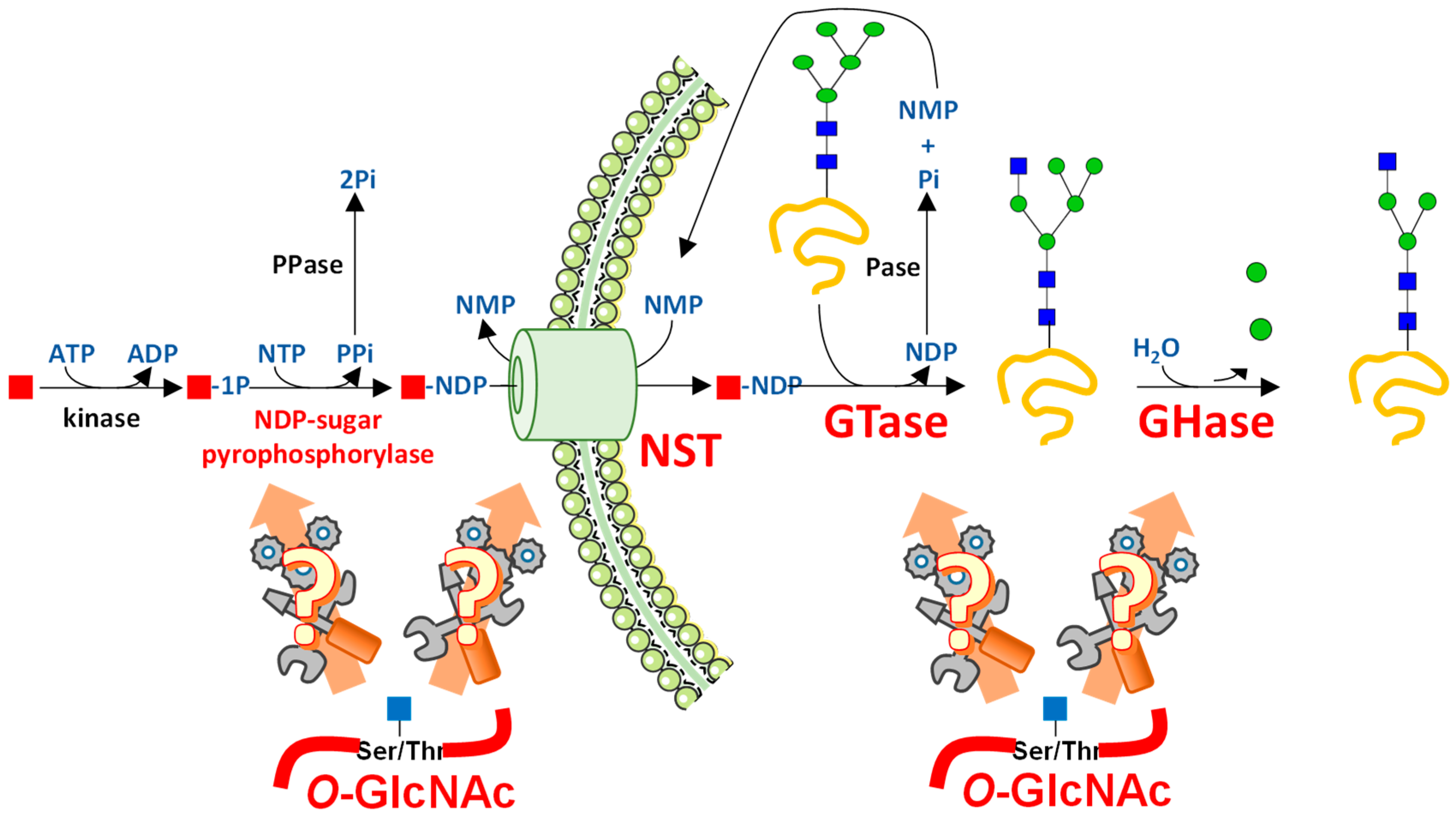
3. UDP-GlcNAc Participates in Many Forms of Glycosylation
3.1. UDP-GlcNAc Is Produced by the HBP and Can Be Converted into UDP-GalNAc and CMP-NeuAc
3.2. UDP-GlcNAc Is a Single Substrate of Many Suitors
3.2.1. N-Glycosylation
3.2.2. O-Glycosylation
3.2.3. Lewis Antigens
3.2.4. Glycosaminoglycans
3.2.5. Glycosphingolipids and Glypiation
4. Interfering with O-GlcNAc Cycling Disrupts Production of Nucleotide Sugars through Expression of HBP Enzymes
5. O-GlcNAcylation Widely and Finely Orchestrates Gene Expression
6. O-GlcNAcylation Regulates Protein Expression
6.1. O-GlcNAc Assists Protein Translation
6.2. O-GlcNAc May Interfere with Protein Degradation through Different Pathways
7. O-GlcNAcylation Orchestrates Vesicular Trafficking and Therefore May Redistribute Glycosylation Enzymes
8. Future Directions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Varki, A.; Sharon, N. Historical Background and Overview. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Corfield, A.P.; Berry, M. Glycan Variation and Evolution in the Eukaryotes. Trends Biochem. Sci. 2015, 40, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Gabius, H.-J. The Sugar Code: Why Glycans Are so Important. Biosystems 2018, 164, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Very, N.; Lefebvre, T.; El Yazidi-Belkoura, I. Drug Resistance Related to Aberrant Glycosylation in Colorectal Cancer. Oncotarget 2018, 9, 1380–1402. [Google Scholar] [CrossRef] [PubMed]
- Vercoutter-Edouart, A.-S.; El Yazidi-Belkoura, I.; Guinez, C.; Baldini, S.; Leturcq, M.; Mortuaire, M.; Mir, A.-M.; Steenackers, A.; Dehennaut, V.; Pierce, A.; et al. Detection and Identification of O-GlcNAcylated Proteins by Proteomic Approaches. Proteomics 2015, 15, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Eustice, M.; Bond, M.R.; Hanover, J.A. O-GlcNAc Cycling and the Regulation of Nucleocytoplasmic Dynamics. Biochem. Soc. Trans. 2017, 45, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging Mechanisms and Functions. Nat. Rev. Mol. Cell. Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed]
- van der Laarse, S.A.M.; Leney, A.C.; Heck, A.J.R. Crosstalk between Phosphorylation and O-GlcNAcylation: Friend or Foe. FEBS J. 2018, 285, 3152–3167. [Google Scholar] [CrossRef] [PubMed]
- Akan, I.; Stichelen, S.O.-V.; Bond, M.R.; Hanover, J.A. Nutrient-Driven O-GlcNAc in Proteostasis and Neurodegeneration. J. Neurochem. 2018, 144, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Gil, M.; Pierce, A.; Perez-Cervera, Y.; Zenteno, E.; Lefebvre, T. OGT: A Short Overview of an Enzyme Standing out from Usual Glycosyltransferases. Biochem. Soc. Trans. 2017, 45, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Sakaidani, Y.; Nomura, T.; Matsuura, A.; Ito, M.; Suzuki, E.; Murakami, K.; Nadano, D.; Matsuda, T.; Furukawa, K.; Okajima, T. O-Linked-N-Acetylglucosamine on Extracellular Protein Domains Mediates Epithelial Cell-Matrix Interactions. Nat. Commun. 2011, 2, 583. [Google Scholar] [CrossRef] [PubMed]
- Nagnan-Le Meillour, P.; Vercoutter-Edouart, A.-S.; Hilliou, F.; Le Danvic, C.; Lévy, F. Proteomic Analysis of Pig (Sus scrofa) Olfactory Soluble Proteome Reveals O-Linked-N-Acetylglucosaminylation of Secreted Odorant-Binding Proteins. Front. Endocrinol. 2014, 5, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M.; Senoo, Y.; Ikeda, K.; Takeuchi, H.; Okajima, T. Structural Divergence in O-GlcNAc Glycans Displayed on Epidermal Growth Factor-like Repeats of Mammalian Notch1. Molecules 2018, 23, 1745. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, T.; Issad, T. 30 Years Old: O-GlcNAc Reaches the Age of Reason—Regulation of Cell Signaling and Metabolism by O-GlcNAcylation. Front. Endocrinol. 2015, 6, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, T.; Dehennaut, V.; Guinez, C.; Olivier, S.; Drougat, L.; Mir, A.-M.; Mortuaire, M.; Vercoutter-Edouart, A.-S.; Michalski, J.-C. Dysregulation of the Nutrient/Stress Sensor O-GlcNAcylation Is Involved in the Etiology of Cardiovascular Disorders, Type-2 Diabetes and Alzheimer’s Disease. Biochim. Biophys. Acta 2010, 1800, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H.; Kinoshita, T.; Varki, A. Glycans in Acquired Human Diseases. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Varki, A.; Kannagi, R.; Toole, B.; Stanley, P. Glycosylation Changes in Cancer. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Ghosh, S.K.; Bond, M.R.; Love, D.C.; Ashwell, G.G.; Krause, M.W.; Hanover, J.A. Disruption of O-GlcNAc Cycling in C. Elegans Perturbs Nucleotide Sugar Pools and Complex Glycans. Front. Endocrinol. 2014, 5, 197. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.B.; Hart, G.W. New Insights: A Role for O-GlcNAcylation in Diabetic Complications. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Miyazaki, J.; Hart, G.W. The Transcription Factor PDX-1 Is Post-Translationally Modified by O-Linked N-Acetylglucosamine and This Modification Is Correlated with Its DNA Binding Activity and Insulin Secretion in Min6 Beta-Cells. Arch. Biochem. Biophys. 2003, 415, 155–163. [Google Scholar] [CrossRef]
- Wellen, K.E.; Lu, C.; Mancuso, A.; Lemons, J.M.S.; Ryczko, M.; Dennis, J.W.; Rabinowitz, J.D.; Coller, H.A.; Thompson, C.B. The Hexosamine Biosynthetic Pathway Couples Growth Factor-Induced Glutamine Uptake to Glucose Metabolism. Genes Dev. 2010, 24, 2784–2799. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, M.; Barzilai, N.; Liu, R.; Hu, M.; Chen, W.; Rossetti, L. Role of the Glucosamine Pathway in Fat-Induced Insulin Resistance. J. Clin. Investig. 1997, 99, 2173–2182. [Google Scholar] [CrossRef] [PubMed]
- Guinez, C.; Mir, A.-M.; Leroy, Y.; Cacan, R.; Michalski, J.-C.; Lefebvre, T. Hsp70-GlcNAc-Binding Activity Is Released by Stress, Proteasome Inhibition, and Protein Misfolding. Biochem. Biophys. Res. Commun. 2007, 361, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Haltiwanger, R.S.; Blomberg, M.A.; Hart, G.W. Glycosylation of Nuclear and Cytoplasmic Proteins. Purification and Characterization of a Uridine Diphospho-N-Acetylglucosamine:Polypeptide Beta-N-Acetylglucosaminyltransferase. J. Biol. Chem. 1992, 267, 9005–9013. [Google Scholar] [PubMed]
- Frey, P.A.; Hegeman, A.D. Chemical and Stereochemical Actions of UDP–Galactose 4-Epimerase. Acc. Chem. Res. 2013, 46, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-C.; Huang, C.-H.; Lai, S.-J.; Yang, C.S.; Hsiao, T.-H.; Lin, C.-H.; Fu, P.-K.; Ko, T.-P.; Chen, Y. Mechanism and Inhibition of Human UDP-GlcNAc 2-Epimerase, the Key Enzyme in Sialic Acid Biosynthesis. Sci. Rep. 2016, 6, 23274. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Intracellular Functions of N-Linked Glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef] [PubMed]
- Ryczko, M.C.; Pawling, J.; Chen, R.; Abdel Rahman, A.M.; Yau, K.; Copeland, J.K.; Zhang, C.; Surendra, A.; Guttman, D.S.; Figeys, D.; et al. Metabolic Reprogramming by Hexosamine Biosynthetic and Golgi N-Glycan Branching Pathways. Sci. Rep. 2016, 6, 23043. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Togayachi, A.; Sakai, T.; Iwai, T.; Hiruma, T.; Sato, T.; Okubo, R.; Inaba, N.; Kudo, T.; Gotoh, M.; et al. A Novel Beta1,3-N-Acetylglucosaminyltransferase (Beta3Gn-T8), Which Synthesizes Poly-N-Acetyllactosamine, Is Dramatically Upregulated in Colon Cancer. FEBS Lett. 2005, 579, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Bard, F.; Chia, J. Cracking the Glycome Encoder: Signaling, Trafficking, and Glycosylation. Trends Cell. Biol. 2016, 26, 379–388. [Google Scholar] [CrossRef] [PubMed]
- An, G.; Wei, B.; Xia, B.; McDaniel, J.M.; Ju, T.; Cummings, R.D.; Braun, J.; Xia, L. Increased Susceptibility to Colitis and Colorectal Tumors in Mice Lacking Core 3–Derived O-Glycans. J. Exp. Med. 2007, 204, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Hascall, V.C.; Wang, A.; Tammi, M.; Oikari, S.; Tammi, R.; Passi, A.; Vigetti, D.; Hanson, R.W.; Hart, G.W. The Dynamic Metabolism of Hyaluronan Regulates the Cytosolic Concentration of UDP-GlcNAc. Matrix Biol. J. Int. Soc. Matrix Biol. 2014, 35, 14–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigetti, D.; Deleonibus, S.; Moretto, P.; Karousou, E.; Viola, M.; Bartolini, B.; Hascall, V.C.; Tammi, M.; De Luca, G.; Passi, A. Role of UDP-N-Acetylglucosamine (GlcNAc) and O-GlcNAcylation of Hyaluronan Synthase 2 in the Control of Chondroitin Sulfate and Hyaluronan Synthesis. J. Biol. Chem. 2012, 287, 35544–35555. [Google Scholar] [CrossRef] [PubMed]
- Deen, A.J.; Arasu, U.T.; Pasonen-Seppänen, S.; Hassinen, A.; Takabe, P.; Wojciechowski, S.; Kärnä, R.; Rilla, K.; Kellokumpu, S.; Tammi, R.; et al. UDP-Sugar Substrates of HAS3 Regulate Its O-GlcNAcylation, Intracellular Traffic, Extracellular Shedding and Correlate with Melanoma Progression. Cell. Mol. Life Sci. CMLS 2016, 73, 3183–3204. [Google Scholar] [CrossRef] [PubMed]
- Vigetti, D.; Deleonibus, S.; Moretto, P.; Bowen, T.; Fischer, J.W.; Grandoch, M.; Oberhuber, A.; Love, D.C.; Hanover, J.A.; Cinquetti, R.; et al. Natural Antisense Transcript for Hyaluronan Synthase 2 (HAS2-AS1) Induces Transcription of HAS2 via Protein O-GlcNAcylation. J. Biol. Chem. 2014, 289, 28816–28826. [Google Scholar] [CrossRef] [PubMed]
- Pummill, P.E.; DeAngelis, P.L. Evaluation of Critical Structural Elements of UDP-Sugar Substrates and Certain Cysteine Residues of a Vertebrate Hyaluronan Synthase. J. Biol. Chem. 2002, 277, 21610–21616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreuger, J.; Kjellén, L. Heparan Sulfate Biosynthesis. J. Histochem. Cytochem. 2012, 60, 898–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, M.; Kinoshita, T. Structural Remodeling of GPI Anchors during Biosynthesis and after Attachment to Proteins. FEBS Lett. 2010, 584, 1670–1677. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Martinez, M.; Sengupta, S.; Lee, A.; Wu, X.; Chaerkady, R.; Chatterjee, A.; O’Meally, R.N.; Cole, R.N.; Pandey, A.; et al. Quantitative Phosphoproteomics Reveals Crosstalk between Phosphorylation and O-GlcNAc in the DNA Damage Response Pathway. Proteomics 2015, 15, 591–607. [Google Scholar] [CrossRef] [PubMed]
- Leturcq, M.; Lefebvre, T.; Vercoutter-Edouart, A.-S. O-GlcNAcylation and Chromatin Remodeling in Mammals: An up-to-Date Overview. Biochem. Soc. Trans. 2017, 45, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Oktaba, K.; Müller, J. Essential Role of the Glycosyltransferase Sxc/Ogt in Polycomb Repression. Science 2009, 325, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, K.; Wang, Z.; Hart, G.W. Beta-N-Acetylglucosamine (O-GlcNAc) Is Part of the Histone Code. Proc. Natl. Acad. Sci. USA 2010, 107, 19915–19920. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Smith, E.; Shilatifard, A. The Language of Histone Crosstalk. Cell 2010, 142, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Bauer, C.; Göbel, K.; Nagaraj, N.; Colantuoni, C.; Wang, M.; Müller, U.; Kremmer, E.; Rottach, A.; Leonhardt, H. Phosphorylation of TET Proteins Is Regulated via O-GlcNAcylation by the O-Linked N-Acetylglucosamine Transferase (OGT). J. Biol. Chem. 2015, 290, 4801–4812. [Google Scholar] [CrossRef] [PubMed]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Méndez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 Regulate GlcNAcylation and H3K4 Methylation through OGT and SET1/COMPASS. EMBO J. 2013, 32, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Vella, P.; Scelfo, A.; Jammula, S.; Chiacchiera, F.; Williams, K.; Cuomo, A.; Roberto, A.; Christensen, J.; Bonaldi, T.; Helin, K.; et al. Tet Proteins Connect the O-Linked N-Acetylglucosamine Transferase Ogt to Chromatin in Embryonic Stem Cells. Mol. Cell. 2013, 49, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, Y.; Bian, C.; Fujiki, R.; Yu, X. TET2 Promotes Histone O-GlcNAcylation during Gene Transcription. Nature 2013, 493, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Phoomak, C.; Silsirivanit, A.; Park, D.; Sawanyawisuth, K.; Vaeteewoottacharn, K.; Wongkham, C.; Lam, E.W.-F.; Pairojkul, C.; Lebrilla, C.B.; Wongkham, S. O-GlcNAcylation Mediates Metastasis of Cholangiocarcinoma through FOXO3 and MAN1A1. Oncogene 2018, 37, 5648–5665. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.A.; Young, L.J.T.; An, H.J.; Kronewitter, S.R.; Kim, J.; Miyamoto, S.; Borowsky, A.D.; Chew, H.K.; Lebrilla, C.B. High-Mannose Glycans Are Elevated during Breast Cancer Progression. Mol. Cell. Proteomics 2011, 10, M110.002717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Qian, Y.; Wu, X.; Zhang, Z.; Liu, X.; Zhao, R.; Zhou, L.; Ruan, Y.; Xu, J.; et al. Discovery of Specific Metastasis-Related N-Glycan Alterations in Epithelial Ovarian Cancer Based on Quantitative Glycomics. PLoS ONE 2014, 9, e87978. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-M.; Hwang, M.P.; Kim, Y.-W.; Kim, K.-J.; Jin, J.M.; Kim, Y.H.; Yang, Y.-H.; Lee, K.H.; Kim, Y.-G. Mass Spectrometry-Based N-Linked Glycomic Profiling as a Means for Tracking Pancreatic Cancer Metastasis. Carbohydr. Res. 2015, 413, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Phoomak, C.; Vaeteewoottacharn, K.; Silsirivanit, A.; Saengboonmee, C.; Seubwai, W.; Sawanyawisuth, K.; Wongkham, C.; Wongkham, S. High Glucose Levels Boost the Aggressiveness of Highly Metastatic Cholangiocarcinoma Cells via O-GlcNAcylation. Sci. Rep. 2017, 7, 43842. [Google Scholar] [CrossRef] [PubMed]
- Datta, B.; Ray, M.K.; Chakrabarti, D.; Wylie, D.E.; Gupta, N.K. Glycosylation of Eukaryotic Peptide Chain Initiation Factor 2 (EIF-2)-Associated 67-KDa Polypeptide (P67) and Its Possible Role in the Inhibition of EIF-2 Kinase-Catalyzed Phosphorylation of the EIF-2 Alpha-Subunit. J. Biol. Chem. 1989, 264, 20620–20624. [Google Scholar] [PubMed]
- Datta, B.; Datta, R.; Ghosh, A.; Majumdar, A. Eukaryotic Initiation Factor 2-Associated Glycoprotein, P67, Shows Differential Effects on the Activity of Certain Kinases during Serum-Starved Conditions. Arch. Biochem. Biophys. 2004, 427, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Zeidan, Q.; Wang, Z.; De Maio, A.; Hart, G.W. O-GlcNAc Cycling Enzymes Associate with the Translational Machinery and Modify Core Ribosomal Proteins. Mol. Biol. Cell. 2010, 21, 1922–1936. [Google Scholar] [CrossRef] [PubMed]
- Shrimal, S.; Cherepanova, N.A.; Gilmore, R. Cotranslational and Posttranslocational N-Glycosylation of Proteins in the Endoplasmic Reticulum. Semin. Cell. Dev. Biol. 2015, 41, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, T.-W.; Cecioni, S.; Eskandari, R.; Zandberg, W.F.; Vocadlo, D.J. O-GlcNAc Occurs Cotranslationally to Stabilize Nascent Polypeptide Chains. Nat. Chem. Biol. 2015, 11, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Very, N.; Steenackers, A.; Dubuquoy, C.; Vermuse, J.; Dubuquoy, L.; Lefebvre, T.; El Yazidi-Belkoura, I. Cross Regulation between MTOR Signaling and O-GlcNAcylation. J. Bioenerg. Biomembr. 2018, 50, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Pak, J.; Jang, I.; Cho, J.W. Inhibition of MTOR Affects Protein Stability of OGT. Biochem. Biophys. Res. Commun. 2014, 453, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Sodi, V.L.; Khaku, S.; Krutilina, R.; Schwab, L.P.; Vocadlo, D.J.; Seagroves, T.N.; Reginato, M.J. MTOR/MYC Axis Regulates O-GlcNAc Transferase Expression and O-GlcNAcylation in Breast Cancer. Mol. Cancer Res. MCR 2015, 13, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Proud, C.G. The MTOR Pathway in the Control of Protein Synthesis. Physiology. 2006, 21, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Valvezan, A.J.; Turner, M.; Belaid, A.; Lam, H.C.; Miller, S.K.; McNamara, M.C.; Baglini, C.; Housden, B.E.; Perrimon, N.; Kwiatkowski, D.J.; et al. MTORC1 Couples Nucleotide Synthesis to Nucleotide Demand Resulting in a Targetable Metabolic Vulnerability. Cancer Cell. 2017, 32, 624–638.e5. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Richard, D.; Laplante, M. The Roles of MTOR Complexes in Lipid Metabolism. Annu. Rev. Nutr. 2015, 35, 321–348. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhao, X.; Liang, L.; Pan, X.; Lv, H.; Zhao, Y. Sialyltransferase ST3GAL6 Mediates the Effect of MicroRNA-26a on Cell Growth, Migration, and Invasion in Hepatocellular Carcinoma through the Protein Kinase B/Mammalian Target of Rapamycin Pathway. Cancer Sci. 2017, 108, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhao, X.; Liang, L.; Wang, G.; Li, Y.; Miao, X.; Zhao, Y. MiR-146a and MiR-146b Promote Proliferation, Migration and Invasion of Follicular Thyroid Carcinoma via Inhibition of ST8SIA4. Oncotarget 2017, 8, 28028–28041. [Google Scholar] [CrossRef] [PubMed]
- Ferris, S.P.; Kodali, V.K.; Kaufman, R.J. Glycoprotein Folding and Quality-Control Mechanisms in Protein-Folding Diseases. Dis. Model. Mech. 2014, 7, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Słomińska-Wojewódzka, M.; Sandvig, K. The Role of Lectin-Carbohydrate Interactions in the Regulation of ER-Associated Protein Degradation. Mol. Basel Switz. 2015, 20, 9816–9846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winchester, B. Lysosomal Metabolism of Glycoproteins. Glycobiology 2005, 15, 1R–15R. [Google Scholar] [CrossRef] [PubMed]
- Seino, J.; Wang, L.; Harada, Y.; Huang, C.; Ishii, K.; Mizushima, N.; Suzuki, T. Basal Autophagy Is Required for the Efficient Catabolism of Sialyloligosaccharides. J. Biol. Chem. 2013, 288, 26898–26907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahie, K.; Zachara, N.E. Molecular Functions of Glycoconjugates in Autophagy. J. Mol. Biol. 2016, 428, 3305–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zachara, N.E.; Hart, G.W. O-GlcNAc Modification: A Nutritional Sensor That Modulates Proteasome Function. Trends Cell. Biol. 2004, 14, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.-B.; Nie, Y.; Yang, X. Regulation of Protein Degradation by O-GlcNAcylation: Crosstalk with Ubiquitination. Mol. Cell. Proteomics MCP 2013, 12, 3489–3497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Su, K.; Yang, X.; Bowe, D.B.; Paterson, A.J.; Kudlow, J.E. O-GlcNAc Modification Is an Endogenous Inhibitor of the Proteasome. Cell 2003, 115, 715–725. [Google Scholar] [CrossRef]
- Guinez, C.; Mir, A.-M.; Dehennaut, V.; Cacan, R.; Harduin-Lepers, A.; Michalski, J.-C.; Lefebvre, T. Protein Ubiquitination Is Modulated by O-GlcNAc Glycosylation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yin, X.; Yang, H.; Xu, Y. Histone Demethylase LSD2 Acts as an E3 Ubiquitin Ligase and Inhibits Cancer Cell Growth through Promoting Proteasomal Degradation of OGT. Mol. Cell. 2015, 58, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, P.K.; Parihar, R.; Dwivedi, V.; Lakhotia, S.C.; Ganesh, S. Decreased O-Linked GlcNAcylation Protects from Cytotoxicity Mediated by Huntingtin Exon1 Protein Fragment. J. Biol. Chem. 2014, 289, 13543–13553. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Liang, Q.; Li, L.; Hu, Z.; Wu, F.; Zhang, P.; Ma, Y.; Zhao, B.; Kovács, A.L.; Zhang, Z.; et al. O-GlcNAc-Modification of SNAP-29 Regulates Autophagosome Maturation. Nat. Cell. Biol. 2014, 16, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Lak, B.; Li, J.; Jokitalo, E.; Wang, Y. GRASP55 Senses Glucose Deprivation through O-GlcNAcylation to Promote Autophagosome-Lysosome Fusion. Dev. Cell. 2018, 45, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y. The Golgi Stacking Protein GORASP2/GRASP55 Serves as an Energy Sensor to Promote Autophagosome Maturation under Glucose Starvation. Autophagy 2018, 14, 1649–1651. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.-B.; Ma, Y.; Torres, S.; Zhang, B.; Feriod, C.; Heck, R.M.; Qian, K.; Fu, M.; Li, X.; Nathanson, M.H.; et al. Calcium-Dependent O-GlcNAc Signaling Drives Liver Autophagy in Adaptation to Starvation. Genes Dev. 2017, 31, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Farmaki, T.; Ponnambalam, S.; Prescott, A.R.; Clausen, H.; Tang, B.L.; Hong, W.; Lucocq, J.M. Forward and Retrograde Trafficking in Mitotic Animal Cells. ER-Golgi Transport Arrest Restricts Protein Export from the ER into COPII-Coated Structures. J. Cell. Sci. 1999, 112, 589–600. [Google Scholar] [PubMed]
- Dudognon, P.; Maeder-Garavaglia, C.; Carpentier, J.-L.; Paccaud, J.-P. Regulation of a COPII Component by Cytosolic O-Glycosylation during Mitosis. FEBS Lett. 2004, 561, 44–50. [Google Scholar] [CrossRef]
- Cox, N.J.; Unlu, G.; Bisnett, B.J.; Meister, T.R.; Condon, B.M.; Luo, P.M.; Smith, T.J.; Hanna, M.; Chhetri, A.; Soderblom, E.J.; et al. Dynamic Glycosylation Governs the Vertebrate COPII Protein Trafficking Pathway. Biochemistry 2018, 57, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Gurel, P.S.; Hatch, A.L.; Higgs, H.N. Connecting the Cytoskeleton to the Endoplasmic Reticulum and Golgi. Curr. Biol. CB 2014, 24, R660–R672. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, A.; Olivier-Van Stichelen, S.; Baldini, S.F.; Dehennaut, V.; Toillon, R.-A.; Le Bourhis, X.; El Yazidi-Belkoura, I.; Lefebvre, T. Silencing the Nucleocytoplasmic O-GlcNAc Transferase Reduces Proliferation, Adhesion, and Migration of Cancer and Fetal Human Colon Cell Lines. Front. Endocrinol. 2016, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Pan, Q.; Sun, D.; Chen, W.; Shen, A.; Huang, M.; Ding, J.; Geng, M. O-GlcNAcylation of Cofilin Promotes Breast Cancer Cell Invasion. J. Biol. Chem. 2013, 288, 36418–36425. [Google Scholar] [CrossRef] [PubMed]
- Hedou, J.; Cieniewski-Bernard, C.; Leroy, Y.; Michalski, J.-C.; Mounier, Y.; Bastide, B. O-Linked N-Acetylglucosaminylation Is Involved in the Ca2+ Activation Properties of Rat Skeletal Muscle. J. Biol. Chem. 2007, 282, 10360–10369. [Google Scholar] [CrossRef] [PubMed]
- Dehennaut, V.; Slomianny, M.-C.; Page, A.; Vercoutter-Edouart, A.-S.; Jessus, C.; Michalski, J.-C.; Vilain, J.-P.; Bodart, J.-F.; Lefebvre, T. Identification of Structural and Functional O-Linked N-Acetylglucosamine-Bearing Proteins in Xenopus Laevis Oocyte. Mol. Cell. Proteomics 2008, 7, 2229–2245. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Kang, J.G.; Park, S.Y.; Lee, J.; Oh, Y.J.; Cho, J.W. O-GlcNAcylation of Tubulin Inhibits Its Polymerization. Amino Acids 2011, 40, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Zhu, W.; Anderson, R.A.; Leber, B.; Andrews, D.W. Multiple Post-Translational Modifications Regulate E-Cadherin Transport during Apoptosis. J. Cell Sci. 2012, 125, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liang, Y.; Xu, Z.; Wang, L.; Zhou, F.; Li, Z.; Jin, J.; Yang, Y.; Fang, Z.; Hu, Y.; et al. N-Glycosylation Affects the Adhesive Function of E-Cadherin through Modifying the Composition of Adherens Junctions (AJs) in Human Breast Carcinoma Cell Line MDA-MB-435. J. Cell. Biochem. 2008, 104, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.K.; Weidner, D.A.; Dayal, S.; Schwalbe, R.A. Cell Surface N-Glycans Influence the Level of Functional E-Cadherin at the Cell-Cell Border. FEBS Open Bio 2014, 4, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Seruca, R.; Gärtner, F.; Yamaguchi, Y.; Gu, J.; Taniguchi, N.; Reis, C.A. Modulation of E-Cadherin Function and Dysfunction by N-Glycosylation. Cell. Mol. Life Sci. 2011, 68, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Stateva, S.R.; Villalobo, A. O-GlcNAcylation of the Human Epidermal Growth Factor Receptor. Org. Biomol. Chem. 2015, 13, 8196–8204. [Google Scholar] [CrossRef] [PubMed]
- Burén, S.; Gomes, A.L.; Teijeiro, A.; Fawal, M.-A.; Yilmaz, M.; Tummala, K.S.; Perez, M.; Rodriguez-Justo, M.; Campos-Olivas, R.; Megías, D.; et al. Regulation of OGT by URI in Response to Glucose Confers C-MYC-Dependent Survival Mechanisms. Cancer Cell 2016, 30, 290–307. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Wang, S.; Fu, M.; Zhou, J.; Singh, J.P.; Li, M.-D.; Yang, Y.; Zhang, K.; Wu, J.; Nie, Y.; et al. Transcriptional Regulation of O-GlcNAc Homeostasis Is Disrupted in Pancreatic Cancer. J. Biol. Chem. 2018, 293, 13989–14000. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.P.; Gundogdu, M.; Kempers, M.J.E.; Giltay, J.C.; Pfundt, R.; Elferink, M.; Loza, B.F.; Fuijkschot, J.; Ferenbach, A.T.; van Gassen, K.L.I.; et al. Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem. 2017, 292, 12621–12631. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, K.; Niranjan, T.; Selvan, N.; Teo, C.F.; May, M.; Patel, S.; Weatherly, B.; Skinner, C.; Opitz, J.; Carey, J.; et al. Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 2017, 292, 8948–8963. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.J.; Lund, K.C.; Taylor, R.P.; McClain, D.A. Insulin resistance of glycogen synthase mediated by O-linked N-acetylglucosamine. J. Biol. Chem. 2003, 278, 10022–10027. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.J.; Taylor, R.P.; Jones, D.; McClain, D.A. Hyperglycemia and inhibition of glycogen synthase in streptozotocin-treated mice: Role of O-linked N-acetylglucosamine. J. Biol. Chem. 2004, 279, 20636–20642. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Park, S.Y.; Ji, S.; Jang, I.; Park, S.; Kim, H.S.; Kim, S.M.; Yook, J.I.; Park, Y.I.; Roth, J.; et al. O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J. Biol. Chem. 2009, 284, 34777–34784. [Google Scholar] [CrossRef] [PubMed]



| Symbol | Designation | Glycosylation Process | Subcellular Localization | EC Number | CAZy |
|---|---|---|---|---|---|
| OGT | O-linked β-N-acetylglucosaminyltransferase | O-GlcNAcylation | Cyt., nucl., mit. | 2.4.1.255 | GT41 |
| EOGT | EGF domain-specific O-linked β-N-acetylglucosaminyltransferase | Extracellular O-GlcNAcylation | ER | 2.4.1.255 | GT61 |
| DPAGT 1 Alg7 | UDP-GlcNAc:dolichol-P GlcNAc-1-P transferase | N-glycosylation (first step) | ER | 2.7.8.15 | None |
| Alg13/Alg14 | GlcNAc diphosphodolichol N-acetylglucosaminyltransferase | N-glycosylation (second step) | ER | 2.4.1.141 3.4.19.12 | GT1 |
| MGAT1GnT-I GGNT1 | Mannosyl (α-1,3-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase | N-glycosylation (synthesis of hybrid and complex N-glycans) | Medial Golgi apparatus | 2.4.1.101 | GT13 |
| MGAT2 GnT-II | Mannosyl (α-1,6-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase | N-glycosylation (conversion of oligomannoses to complex N-glycans) | Golgi apparatus | 2.4.1.143 | GT16 |
| MGAT3 GnT-III GGNT3 | Mannosyl (β-1,4-)-glycoprotein β-1,4-N-acetylglucosaminyltransferase | N-glycosylation (bisecting GlcNAc) | Medial-trans Golgi apparatus | 2.4.1.144 | GT17 |
| MGAT4 GnT-IV | Mannosyl (α-1,3-)-glycoprotein β-1,4-N-Acetylglucosaminyltransferase | N-glycosylation (synthesis of tri- and tetra-antennary N-glycans) | Golgi apparatus | 2.4.1.145 | GT54 |
| MGAT5 GnT-V GGNT5 | Mannosyl (α-1,6-)-glycoprotein β-1,6-N-acetylglucosaminyltransferase | N-glycosylation (initiation of β-1,6-branched structures) | Medial-trans Golgi apparatus | 2.4.1.155 | GT18 |
| B3GNT8 | UDP-GlcNAc: β-Gal β-1,3-N-acetylglucosaminyltransferase 8 | N-glycosylation | Golgi apparatus | 2.4.1.- | - |
| C2GnT GCNT1 | core 2 β-1,6-N-acetylglucosaminyltransferase | Mucin-type O-glycosylation (synthesis of core 2) | Golgi apparatus | 2.4.1.102 | GT14 |
| C3GnT B3GNT6 | core 3 β-1,6-N-acetylglucosaminyltransferase | Mucin-type O-glycosylation (synthesis of core 3) | Golgi apparatus | 2.4.1.149 | GT31 |
| C4GnT GCNT3 | Core 2/Core 4 β-1,6-N-acetylglucosaminyltransferase | Mucin-type O-glycosylation (synthesis of cores 2 & 4) | Golgi apparatus | 2.4.1.102 | GT14 |
| HAS1-3 | Hyaluronic acid synthase 1–3 | Hyaluronic acid synthesis | Plasma membrane (Cyt. face) | 2.4.1.212 | GT2 |
| EXT1 | Exostosin like glycosyltransferase 1 | Heparin and heparan sulfate | ER | 2.4.1.224 | GT47 |
| EXT2 | Exostosin like glycosyltransferase 2 | Heparan sulfate | ER and Golgi apparatus | 2.4.1.224 2.4.1.225 | GT47 GT64 |
| EXT3 | Exostosin like glycosyltransferase 3 | Heparin and heparan sulfate | ER and Golgi apparatus | 2.4.1.223 | GT47 |
| B3GNT5 | UDP-GlcNAc: β-Gal β-1,3-N-acetylglucosaminyltransferase 5 | Glycolipids (lacto and neolacto-series; crucial for Lewis X epitope) | Golgi apparatus | 2.4.1.206 | GT31 |
| B3GNT8 | UDP-GlcNAc: β-Gal β-1,3-N-acetylglucosaminyltransferase 8 | N-glycosylation (elongation of branched structures) | Golgi apparatus | 2.4.1.149 | GT31 |
| PIG-A/C/H/P/Q/Y | Phosphatidylinositol N-acetylglucosaminyltransferase subunits A, C, H, P, Q, and Y | GPI-anchors (synthesis of GlcNAc-phosphatidylinositol) | ER membrane (Cyt. face) | 2.4.1.198 | GT4 |
| Symbol | Designation | Glycosylation Process | Subcellular Localization | EC Number | CAZy |
|---|---|---|---|---|---|
| Upregulated | |||||
| GLB1 | β-galactosidase (beta 1) | Active on gangliosides, glycoproteins and GAG | Lysosome | 3.2.1.23 | GH35 |
| FUT10 | Fucosyltransferase 10 (α-1,3 fucosyltransferase) | Synthesis of Lewis X on N-glycans | Golgi apparatus | 2.4.1.65 | GT10 |
| FUT8 | Fucosyltransferase 8 (α-1,6 fucosyltransferase) | Active on complex N-type glycans | Golgi apparatus | 2.4.1.68 | GT23 |
| MAN2A1 | α-mannosidase, class 2A, member 1 | Maturation of N-glycans | Golgi apparatus | 3.2.1.114 | GH38 |
| MGAT5 GnT-V GGNT5 | Mannosyl (α-1,6-)-glycoprotein β-1,6-N-acetylglucosaminyltransferase | N-glycosylation (initiation of β-1,6-branched structures) | Medial-trans Golgi apparatus | 2.4.1.155 | GT18 |
| B4GALT6 | UDP-Gal: βGlcNAc β-1,4 GalTase, polypeptide 6 | Glycolipids (synthesis of lactosylceramide) | Medial-trans Golgi apparatus | 2.4.1.274 | GT7 |
| B4GALT7 | xylosylprotein β-1,4-galactosyltransferase, polypeptide 7 | Proteoglycans | Golgi apparatus | 2.4.1.133 | GT7 |
| B3GNT5 | UDP-GlcNAc: β-Gal β-1,3-N-acetylglucosaminyltransferase 5 | Glycolipids (lacto and neolacto-series; crucial for Lewis X epitope) | Golgi apparatus | 2.4.1.206 | GT31 |
| UGGT1 | UDP-Glc glycoprotein GlcTfase 1 | N-glycosylation (glucosylation of unfolded proteins) | ER | 2.4.1.- | GT24 |
| GALNT1 | UDP-N-GalNAc:polypeptide GalNAcTase 1 (GalNAc-T1) | O-glycosylation (mucin-type) | Golgi apparatus | 2.4.1.41 | GT27 |
| GALNT10 | UDP-N-GalNAc:polypeptide GalNAcTase 10 (GalNAc-T10) | O-glycosylation (mucin-type) | Golgi apparatus | 2.4.1.41 | GT27 |
| GALNT12 | UDP-N-GalNAc:polypeptide GalNAcTase 12 (GalNAc-T12) | O-glycosylation (mucin-type) | Golgi apparatus | 2.4.1.41 | GT27 |
| Downregulated | |||||
| Alg14 | Asn-linked glycosylation 14 homolog (S. cerevisiae) | N-glycosylation (second step) | ER | 2.4.1.141 | None |
| B4GALNT4 | β-1,4-N-acetyl-galactosaminyltransferase 4 | N-glycosylation | Golgi apparatus | 2.4.1.244 | GT7 |
| OGT | O-linked β-N-acetylglucosaminyltransferase | O-GlcNAcylation | Cyt., nucl., mit. | 2.4.1.255 | GT41 |
| Process | Reference |
|---|---|
| Experimentally proved | |
| Nucleotide sugars levels | |
| OGT and OGA interfere with UDP-Glc and UDP-HexNAc production | [20] |
| Expression of enzymes of HBP | |
| In ogt-ko animals, mRNAs encoding gfat2, gna-2, and the putative UDP-GlcNAc pyrophosphorylase C36A4.4 are up-regulated | [20] |
| UAP1 and Gnpda1 are upregulated in OGT NULL MEFs | [41] |
| Transcriptional regulation | |
| Transiently OGT-depleted mESCs exhibit either up- or down-regulation of genes involved in N- and O-glycosylations controlled by OGT | [48] |
| OGT regulates high-mannose N-linked glycans: OGT signaling in cholangiocarcinoma cells decreases MAN1A1 expression through a down-regulation of the MAPK-FOXO3 axis | [50] |
| Protein synthesis through mTOR | |
| ST3GAL6 expression correlates with mTOR activation in hepatoma carcinoma cells | [66] |
| ST8SIA4 expression is negatively correlated with mTOR activation in follicular thyroid cancer cells | [67] |
| Speculative | |
| Nucleotide sugar levels | |
| Competition for UDP-GlcNAc between OGT and other GTase (HAS, EOGT, reticular, and golgian GlcNAc transferases) | |
| Transcriptional regulation | |
| Transcriptional regulation of genes involved in glycosylation processes including nucleotide sugar transporters, GTases and GHases | |
| Protein synthesis | |
| Translation of glycosylation actors: Protection of eIF-2 by binding to O-GlcNAc forms of p67 | [55,56] |
| OGT and OGA are partners of ribosomes; several ribosomal proteins are O-GlcNAcylated (e.g., RPS6) | [57] |
| Stabilization of nascent proteins by O-GlcNAcylation to prevent premature degradation | [59] |
| mTOR pathway is controlled by O-GlcNAcylation: Expression of glycosylation enzymes may be under the control of mTOR | [60,61,62] |
| Vesicular traffic | |
| Traffic of vesicular compounds through COPII | [85] |
| Through SEC23A, SEC24C, SEC31A, and TFG | [84,86] |
| Through the cytoskeleton | [87,88,89,90,91] |
| Through small G-proteins (Rab) | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biwi, J.; Biot, C.; Guerardel, Y.; Vercoutter-Edouart, A.-S.; Lefebvre, T. The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations. Molecules 2018, 23, 2858. https://doi.org/10.3390/molecules23112858
Biwi J, Biot C, Guerardel Y, Vercoutter-Edouart A-S, Lefebvre T. The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations. Molecules. 2018; 23(11):2858. https://doi.org/10.3390/molecules23112858
Chicago/Turabian StyleBiwi, James, Christophe Biot, Yann Guerardel, Anne-Sophie Vercoutter-Edouart, and Tony Lefebvre. 2018. "The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations" Molecules 23, no. 11: 2858. https://doi.org/10.3390/molecules23112858
APA StyleBiwi, J., Biot, C., Guerardel, Y., Vercoutter-Edouart, A.-S., & Lefebvre, T. (2018). The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations. Molecules, 23(11), 2858. https://doi.org/10.3390/molecules23112858






