Biochemical and Functional Characterization of Anthocyanidin Reductase (ANR) from Mangifera indica L.
Abstract
:1. Introduction
2. Results
2.1. Molecular Cloning of ANR Homolog from Hongguifei Mango
2.2. Functional Expression in E. coli and In Vitro Biochemical Characterization
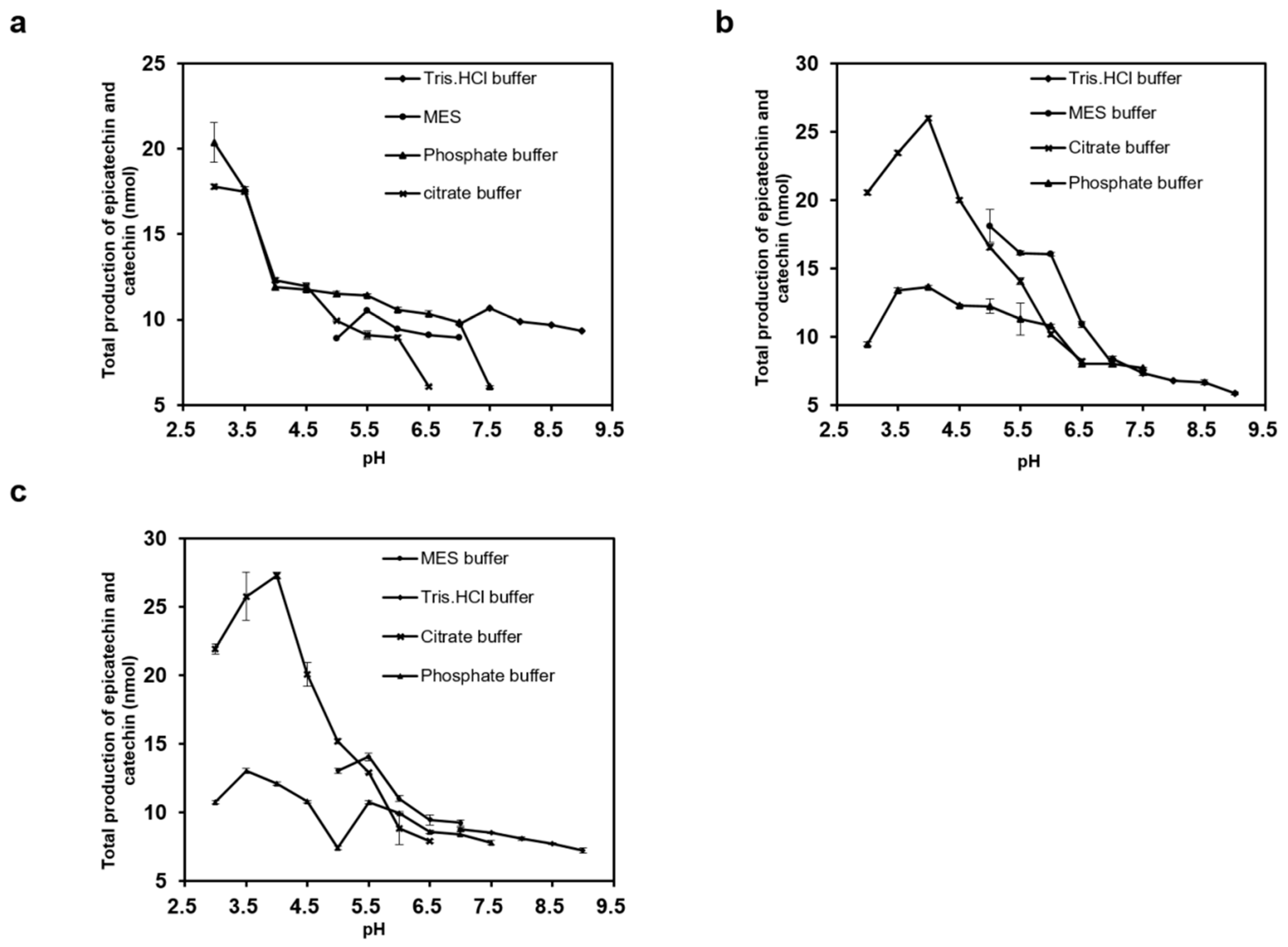
2.3. Effects of Temperature, pH, and NaCl on ANR Activities
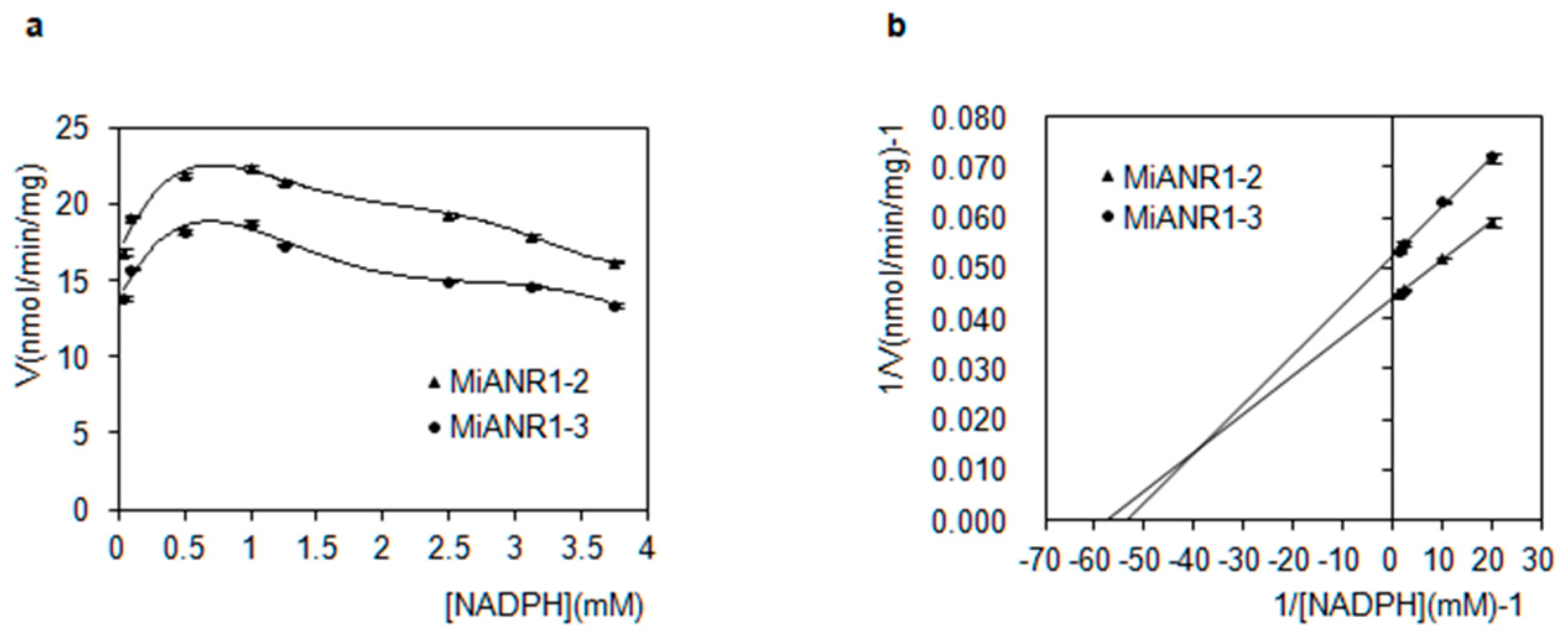
2.4. Kinetics of MiANRs with Coenzymes, Anthocyanidins and Perlargonidins
2.5. Over-Expression of MiANR1-1, 1-2 and 1-3 in AtANR Mutant (Banyuls) Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. cDNA Cloning of ANR from Pulp of Hongguifei Mango by RACE
4.3. Expression of Recombinant MiANR cDNAs in E. coli
4.4. Purification of Recombinant MiANRs
4.5. Enzymatic Assay
4.6. Determination of Kinetic Parameters for Recombinant MiANRs
4.7. UHPLC-UV-MS and Ultraviolet–Visible Spectrometry Analyses
4.8. Over-Expression of MiANR in the Banuyls Mutant of Arabidopsis thaliana
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ulusoy, S.; Ozkan, G.; Yucesan, F.B.; Ersöz, Ş.; Orem, A.; Alkanat, M.; Yuluğ, E.; Kaynar, K.; Al, S. Anti-apoptotic and anti-oxidant effects of grape seed proanthocyanidin extract in preventing cyclosporine A-induced nephropathy. Nephrology 2012, 17, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Sobeh, M.; Mahmoud, M.F.; Abdelfattah, M.A.O.; Cheng, H.; El-Shazly, A.M.; Wink, M. A proanthocyanidin-rich extract from Cassia abbreviata exhibits antioxidant and hepatoprotective activities in vivo. J. Ethnopharmacol. 2018, 213, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.-J.; Liu, H.-B.; Yu, Y.; Liang, L.; Xu, R.; Liang, C.; Tang, J.-S.; Yao, X.-S. Anticancer activities of proanthocyanidins from the plant Urceola huaitingii and their synergistic effects in combination with chemotherapeutics. Fitoterapia 2016, 112, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Padumadasa, C.; Dharmadana, D.; Abeysekera, A.; Thammitiyagodage, M. In vitro antioxidant, anti-inflammatory and anticancer activities of ethyl acetate soluble proanthocyanidins of the inflorescence of Cocos nucifera L. BMC Complement. Altern. Med. 2016, 16, 345. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.E.; Frederiksen, H.; Struntze Krogholm, K.; Poulsen, L. Dietary proanthocyanidins: Occurrence, dietary intake, bioavailability, and protection against cardiovascular disease. Mol. Nutr. Food Res. 2005, 49, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Vaid, M.; Singh, T.; Prasad, R.; Elmets, C.A.; Xu, H.; Katiyar, S.K. Bioactive grape proanthocyanidins enhance immune reactivity in UV-irradiated skin through functional activation of dendritic cells in mice. Cancer Prev. Res. 2013, 6, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Dinh, J.; Angeloni, J.T.; Pederson, D.B.; Wang, X.; Cao, M.; Dong, Y. Cranberry extract standardized for proanthocyanidins promotes the immune response of Caenorhabditis elegans to Vibrio cholerae through the p38 MAPK pathway and HSF-1. PLoS ONE 2014, 9, e103290. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Ma, D.; Constabel, C.P. The MYB182 Protein Down-Regulates Proanthocyanidin and Anthocyanin Biosynthesis in Poplar by Repressing Both Structural and Regulatory Flavonoid Genes. Plant Physiol. 2015, 167, 693–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeynayake, S.W.; Panter, S.; Chapman, R.; Webster, T.; Rochfort, S.; Mouradov, A.; Spangenberg, G. Biosynthesis of Proanthocyanidins in White Clover Flowers: Cross Talk within the Flavonoid Pathway. Plant Physiol. 2012, 158, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Hisano, T.; Yasui, Y.; Mori, M.; Walker, A.R.; Morishita, T.; Katsu, K. Isolation and characterization of genes encoding leucoanthocyanidin reductase (FeLAR) and anthocyanidin reductase (FeANR) in buckwheat (Fagopyrum esculentum). J. Plant Physiol. 2016, 205, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Abeysinghe, I.S.B.; He, J.; He, X.; Huhman, D.; Mewan, K.M.; Sumner, L.W.; Yun, J.; Dixon, R.A. Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 2013, 161, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Kovinich, N.; Saleem, A.; Arnason, J.T.; Miki, B. Identification of Two Anthocyanidin Reductase Genes and Three Red-Brown Soybean Accessions with Reduced Anthocyanidin Reductase 1 mRNA, Activity, and Seed Coat Proanthocyanidin Amounts. J. Agric. Food Chem. 2012, 60, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin-Wang, K.; Liao, L.; Gu, C.; Lu, Z.; Allan, A.C.; Han, Y. Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Front. Plant Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Vimolmangkang, S.; Wei, G.; Zhou, H.; Korban, S.S.; Han, Y. Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 2015, 6, 243. [Google Scholar] [CrossRef] [PubMed]
- Ayres, M.; Clausen, T.; MacLean, S.; Redman, A.; Relchardt, P. Diversity of structure and antiherbivore activity in condensed tannins. Ecology 1997, 78, 1696–1712. [Google Scholar] [CrossRef]
- Mellway, R.D.; Tran, L.T.; Prouse, M.B.; Campbell, M.M.; Constabel, C.P. The Wound-, Pathogen-, and Ultraviolet B-Responsive MYB134 Gene Encodes an R2R3 MYB Transcription Factor That Regulates Proanthocyanidin Synthesis in Poplar. Plant Physiol. 2009, 150, 924–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Yao, G.; Yue, W.; Zhang, S.; Wu, J. Transcriptome profiling reveals differential gene expression in proanthocyanidin biosynthesis associated with red/green skin color mutant of pear (Pyrus communis L.). Front. Plant Sci. 2015, 6, 795. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. PPB 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanner, G.J.; Francki, K.T.; Abrahams, S.; Watson, J.M.; Larkin, P.J.; Ashton, A.R. Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 2003, 278, 31647–31656. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, Y.; Yuan, L.; Lu, W.; Yang, L.; Karim, A.; Luo, K. Isolation and characterization of cDNAs encoding leucoanthocyanidin reductase and anthocyanidin reductase from Populus trichocarpa. PLoS ONE 2013, 8, e64664. [Google Scholar] [CrossRef] [PubMed]
- Paolocci, F.; Robbins, M.P.; Madeo, L.; Arcioni, S.; Martens, S.; Damiani, F. Ectopic Expression of a Basic Helix-Loop-Helix Gene Transactivates Parallel Pathways of Proanthocyanidin Biosynthesis. Structure, Expression Analysis, and Genetic Control of Leucoanthocyanidin 4-Reductase and Anthocyanidin Reductase Genes in Lotus cornicu. Plant Physiol. 2006, 143, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Peng, Q.-Z.; Li, K.-G.; Xie, D.-Y. Molecular cloning and functional characterization of the anthocyanidin reductase gene from Vitis bellula. Planta 2014, 240, 381–398. [Google Scholar] [CrossRef] [PubMed]
- Mandave, P.C.; Kuvalekar, A.A.; Mantri, N.L.; Autal Islam, M.; Ranjekar, P.K. Cloning, expression and molecular modeling of the anthocyanidin reductase (FaANR) gene during strawberry fruit development. Fruits 2017, 72, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Ikegami, A.; Eguchi, S.; Kitajima, A.; Inoue, K.; Yonemori, K. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 2007, 172, 1037–1047. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Gesell, A.; Yoshida, K.; Tran, L.T.; Constabel, C.P. Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 2014, 240, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Gagné, S.; Lacampagne, S.; Claisse, O.; Gény, L. Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol. Biochem. 2009, 47, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P. Effect of potassium nutrition on mineral composition of mango leaf and ripe fruit. Indian J. Hortic. 1999, 56, 238–241. [Google Scholar]
- Vergara-Valencia, N.; Granados-Pérez, E.; Agama-Acevedo, E.; Tovar, J.; Ruales, J.; Bello-Pérez, L.A. Fibre concentrate from mango fruit: Characterization, associated antioxidant capacity and application as a bakery product ingredient. LWT Food Sci. Technol. 2007. [Google Scholar] [CrossRef]
- Sudha, M.L.; Indumathi, K.; Sumanth, M.S.; Rajarathnam, S.; Shashirekha, M.N. Mango pulp fibre waste: Characterization and utilization as a bakery product ingredient. J. Food Meas. Charact. 2015. [Google Scholar] [CrossRef]
- Sellamuthu, P.S.; Denoya, G.I.; Sivakumar, D.; Polenta, G.A.; Soundy, P. Comparison of the contents of bioactive compounds and quality parameters in selected mango cultivars. J. Food Qual. 2013. [Google Scholar] [CrossRef]
- Matkowski, A.; Kus, P.; Goralska, E.; Wozniak, D. Mangiferin—A bioactive xanthonoid, not only from mango and not just antioxidant. Mini Rev. Med. Chem. 2013. [Google Scholar] [CrossRef]
- Das, S.; Alam, M.D.N.; Batuta, S.; Roy, N.; Begum, N.A. Exploring Comparative Antioxidant Activity of Some Popular Cultivars of Mangifera indica L., National Fruit of India. Int. J. Fruit Sci. 2015. [Google Scholar] [CrossRef]
- Ma, X.; Wu, H.; Liu, L.; Yao, Q.; Wang, S.; Zhan, R.; Xing, S.; Zhou, Y. Polyphenolic compounds and antioxidant properties in mango fruits. Sci. Hortic. 2011. [Google Scholar] [CrossRef]
- Dorta, E.; González, M.; Lobo, M.G.; Laich, F. Antifungal activity of mango peel and seed extracts against clinically pathogenic and food spoilage yeasts. Nat. Prod. Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dare, A.P.; Tomes, S.; Jones, M.; McGhie, T.K.; Stevenson, D.E.; Johnson, R.A.; Greenwood, D.R.; Hellens, R.P. Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica). Plant J. 2013. [Google Scholar] [CrossRef] [PubMed]
- Massonnet, M.; Fasoli, M.; Tornielli, G.B.; Altieri, M.; Sandri, M.; Zuccolotto, P.; Paci, P.; Gardiman, M.; Zenoni, S.; Pezzotti, M. Ripening Transcriptomic Program in Red and White Grapevine Varieties Correlates with Berry Skin Anthocyanin Accumulation. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L. A key structural gene, AaLDOX, is involved in anthocyanin biosynthesis in all red-fleshed kiwifruit (Actinidia arguta) based on transcriptome analysis. Gene 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Li, G.; Hu, P.; Zhao, X.; Li, L.; Wei, W.; Feng, J.; Zhou, H. Identification of basic/helix-loop-helix transcription factors reveals candidate genes involved in anthocyanin biosynthesis from the strawberry white-flesh mutant. Sci. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, B.; Gao, A.; Chen, Y.; Huang, J.; Dang, Z.; Luo, R. Cloning and Expression Analysis of UFGT Gene from Mango (Mangifera indica). Jiangsu Agric. Sci. 2016, 44, 31–34. [Google Scholar]
- Zhao, Z.; Gao, A.; Huang, J.; Dang, Z.; Luo, R.; Chen, Y. Cloning and Expression Analysis of F3’ H Gene from Mango (Mangifera indica). Hans J. Comput. Biol. 2016, 6, 1–7. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, Z.; Gao, A.; Luo, R. Cloning and Expression Analysis of DFR Gene from Mango (Mangifera indica). Mol. Plant Breed. 2015, 4, 816–821. [Google Scholar]
- Li, X.; Zhao, Z.; Gao, A.; Chen, Y.; Huang, J.; Dang, Z.; Luo, R. Cloning and Expression Analysis of ANR Gene from Mango (Mangifera indica). Jiangsu Agric. Sci. 2017, 45, 22–25. [Google Scholar]
- Luria, N.; Sela, N.; Yaari, M.; Feygenberg, O.; Kobiler, I.; Lers, A.; Prusky, D. De-novo assembly of mango fruit peel transcriptome reveals mechanisms of mango response to hot water treatment. BMC Genom. 2014, 15, 957. [Google Scholar] [CrossRef] [PubMed]
- Gargouri, M.; Manigand, C.; Maugé, C.; Granier, T.; Langlois d’Estaintot, B.; Cala, O.; Pianet, I.; Bathany, K.; Chaudière, J.; Gallois, B. Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-Y.; Sharma, S.B.; Dixon, R.A. Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch. Biochem. Biophys. 2004, 422, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Delseny, M.; Devic, M. BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 1997, 11, 289–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devic, M.; Guilleminot, J.; Debeaujon, I.; Bechtold, N.; Bensaude, E.; Koornneef, M.; Pelletier, G.; Delseny, M. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 1999, 19, 387–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scrutton, N.S.; Berry, A.; Perham, R.N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 1990, 343, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Marohnic, C.C.; Bewley, M.C.; Barber, M.J. Engineering and characterization of a NADPH-utilizing cytochrome b5 reductase. Biochemistry 2003, 42, 11170–11182. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, S.P.; Mathews, C.K. Allosteric regulation of vaccinia virus ribonucleotide reductase, analyzed by simultaneous monitoring of its four activities. J. Biol. Chem. 1998, 273, 29512–29518. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Zhang, L.; Jiang, S.; Yao, Z.; Gao, B.; Lin, J.; Yuan, Y.A.; Wei, D. A computational strategy for altering an enzyme in its cofactor preference to NAD(H) and/or NADP(H). FEBS J. 2015, 282, 2339–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kho, R.; Baker, B.L.; Newman, J.V.; Jack, R.M.; Sem, D.S.; Villar, H.O.; Hansen, M.R. A path from primary protein sequence to ligand recognition. Proteins 2003, 50, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.Y.; Jackson, L.A.; Cooper, J.D.; Ferreira, D.; Paiva, N.L. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 2004. [Google Scholar] [CrossRef] [PubMed]
- Miosic, S.; Thill, J.; Milosevic, M.; Gosch, C.; Pober, S.; Molitor, C.; Ejaz, S.; Rompel, A.; Stich, K.; Halbwirth, H. Dihydroflavonol 4-reductase genes encode enzymes with contrasting substrate specificity and show divergent gene expression profiles in Fragaria species. PLoS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Karanjalker, G.R.; Ravishankar, K.V.; Shivashankara, K.S.; Dinesh, M.R.; Roy, T.K.; Sudhakar Rao, D.V. A Study on the Expression of Genes Involved in Carotenoids and Anthocyanins During Ripening in Fruit Peel of Green, Yellow, and Red Colored Mango Cultivars. Appl. Biochem. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Rymbai, H.; Srivastav, M.; Sharma, R.R.; Patel, C.R.; Singh, A.K. Bio-active compounds in mango (Mangifera indica L.) and their roles in human health and plant defence—A review. J. Hortic. Sci. Biotechnol. 2013, 88, 369–379. [Google Scholar] [CrossRef]
- Ottaviani, J.I.; Momma, T.Y.; Heiss, C.; Kwik-Uribe, C.; Schroeter, H.; Keen, C.L. The stereochemical configuration of flavanols influences the level and metabolism of flavanols in humans and their biological activity in vivo. Free Radic. Biol. Med. 2011, 50, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, Z.; Maximova, S.; Payne, M.J.; Guiltinan, M.J. Proanthocyanidin synthesis in Theobroma cacao: Genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 2013, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liao, L.; Zhou, H.; Wang, L.; Deng, X.; Han, Y. Constitutive activation of an anthocyanin regulatory gene PcMYB10.6 is related to red coloration in purple-foliage plum. PLoS ONE 2015, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.C.; Gosch, C.; Pfeiffer, J.; Halbwirth, H.; Halle, C.; Stich, K.; Forkmann, G. Flavonoid genes of pear (Pyrus communis). Trees 2007, 21, 521–529. [Google Scholar] [CrossRef]
- Peel, G.; Dixon, R. Detection and quantification of engineered proanthocyanidins in transgenic plants. Nat. Prod. Commun. 2007, 2, 1009–1014. [Google Scholar]
- Thies, M.; Fischer, R. New color reaction for microchemical detection and quantitative determination of catechins. Mikrochim. Acta 1971, 1, 9–13. [Google Scholar] [CrossRef]
- Treutter, D. Chemical reaction detection of catechins and proanthocyanidins with 4-dimethylaminocinnamaldehyde. J. Chromatogr. A 1989. [Google Scholar] [CrossRef]
- Hofgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988. [Google Scholar] [CrossRef]
- Logemann, E.; Birkenbihl, R.P.; Ülker, B.; Somssich, I.E. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2006, 2, 16. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |







| Enzyme | Substrates | Cofactors | |
|---|---|---|---|
| Cyanidin | NADPH | ||
| MiANR1-1 | Vmax (nmol/min mg) | 9.11 ± 0.15 | NR |
| Km (μM) | 48.32 ± 3.45 | - | |
| Kcat (min−1) | 958 | - | |
| Kcat/Km (M−1 S−1) | 3.3 × 105 | - | |
| MiANR1-2 | Vmax (nmol/min mg) | 24.15 ± 0.92 | 22.68 ± 0.18 |
| Km (μM) | 116.4 ± 4.42 | 18.14 ± 1.4 | |
| Kcat (min−1) | 2542 | - | |
| Kcat/Km (M−1 S−1) | 3.63 × 105 | - | |
| MiANR1-3 | Vmax (nmol/min mg) | 18.42 ± 1.38 | 18.98 ± 0.1 |
| Km (μM) | 80.43 ± 5.85 | 18.98 ± 1.18 | |
| Kcat (min−1) | 1938 | - | |
| Kcat/Km (M−1 S−1) | 4.01 × 105 | - | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, L.; Wang, M.; Kang, Y.; Azeem, F.; Zhou, Z.; Tuo, D.; María Preciado Rojo, L.; Khan, I.A.; Pan, Z. Biochemical and Functional Characterization of Anthocyanidin Reductase (ANR) from Mangifera indica L. Molecules 2018, 23, 2876. https://doi.org/10.3390/molecules23112876
Tan L, Wang M, Kang Y, Azeem F, Zhou Z, Tuo D, María Preciado Rojo L, Khan IA, Pan Z. Biochemical and Functional Characterization of Anthocyanidin Reductase (ANR) from Mangifera indica L. Molecules. 2018; 23(11):2876. https://doi.org/10.3390/molecules23112876
Chicago/Turabian StyleTan, Lin, Mei Wang, Youfa Kang, Farrukh Azeem, Zhaoxi Zhou, Decai Tuo, Lina María Preciado Rojo, Ikhlas A. Khan, and Zhiqiang Pan. 2018. "Biochemical and Functional Characterization of Anthocyanidin Reductase (ANR) from Mangifera indica L." Molecules 23, no. 11: 2876. https://doi.org/10.3390/molecules23112876





