Selenate Prevents Adipogenesis through Induction of Selenoprotein S and Attenuation of Endoplasmic Reticulum Stress
Abstract
1. Introduction
2. Results
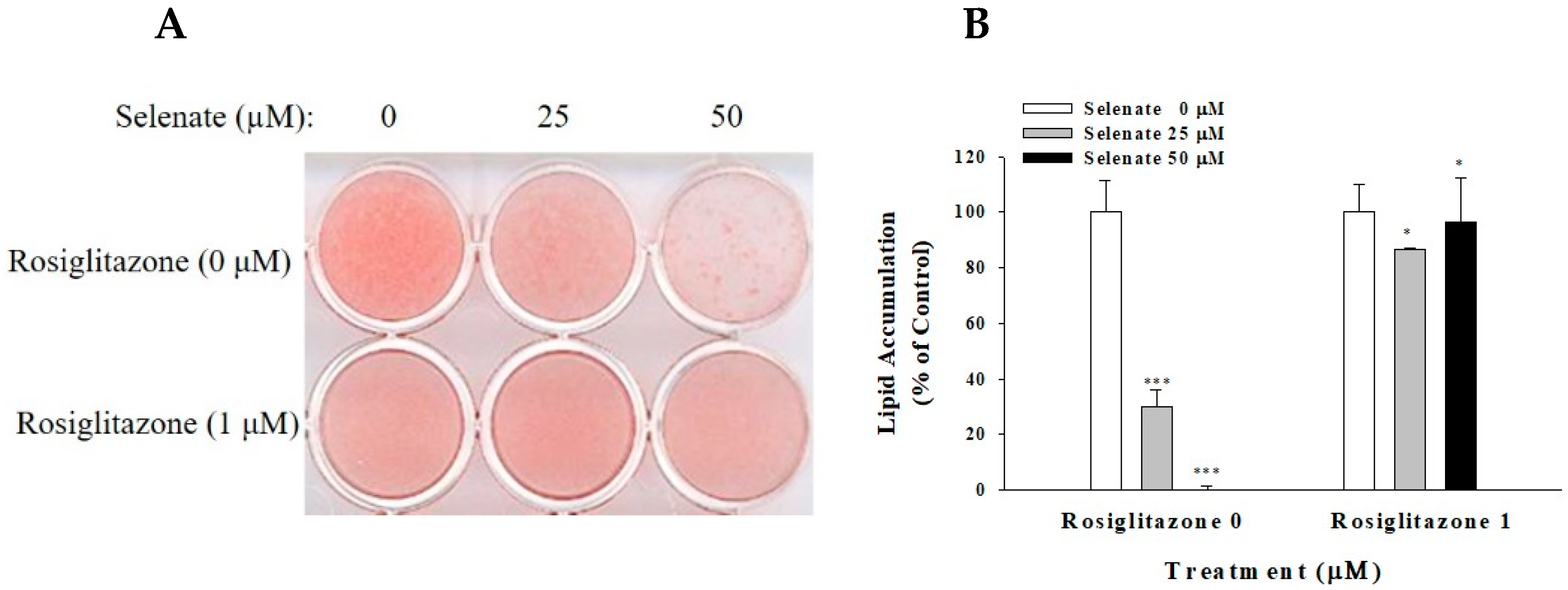
2.1. Selenate Pretreatment Suppresses Adipogenesis in 3T3-L1 Preadipocytes
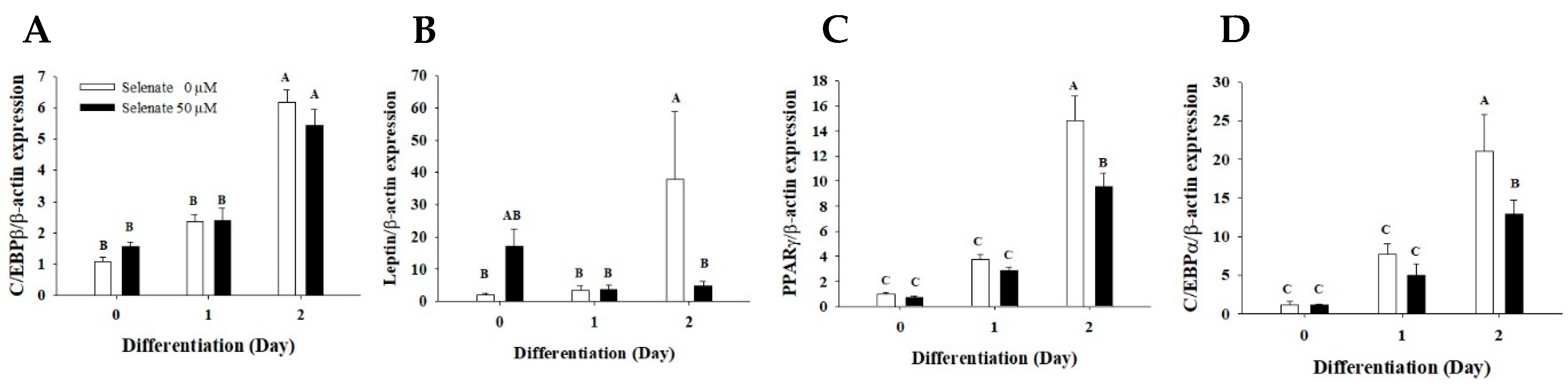
2.2. Selenate Pretreatment Inhibits PPARγ and C/EBPα Gene Expression in the Early Phase of Adipogenesis
2.3. Regulation of SEPS1 Protein by Selenium and Dexamethasone Treatment
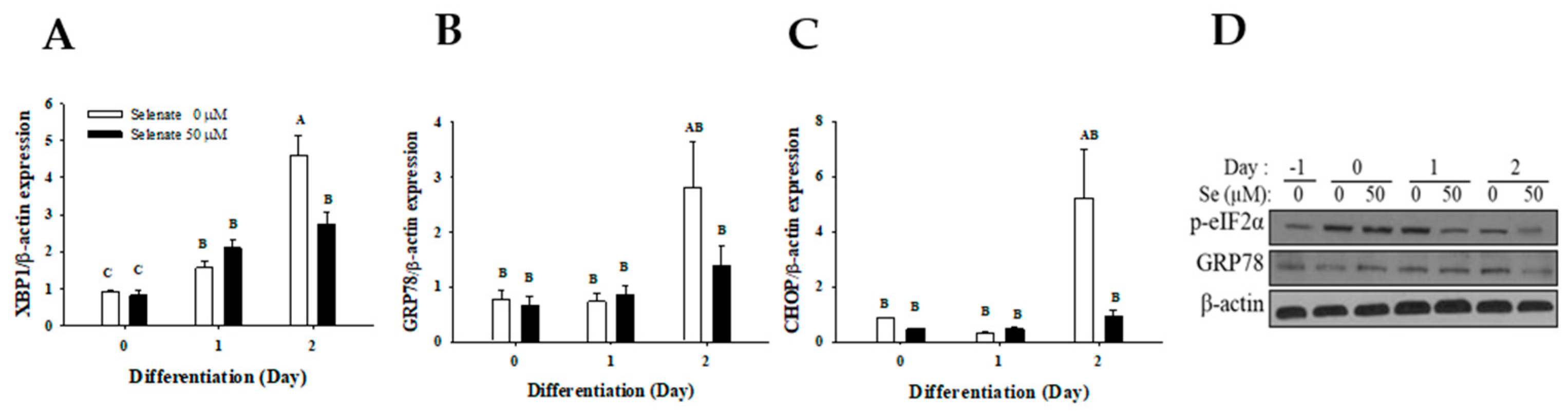
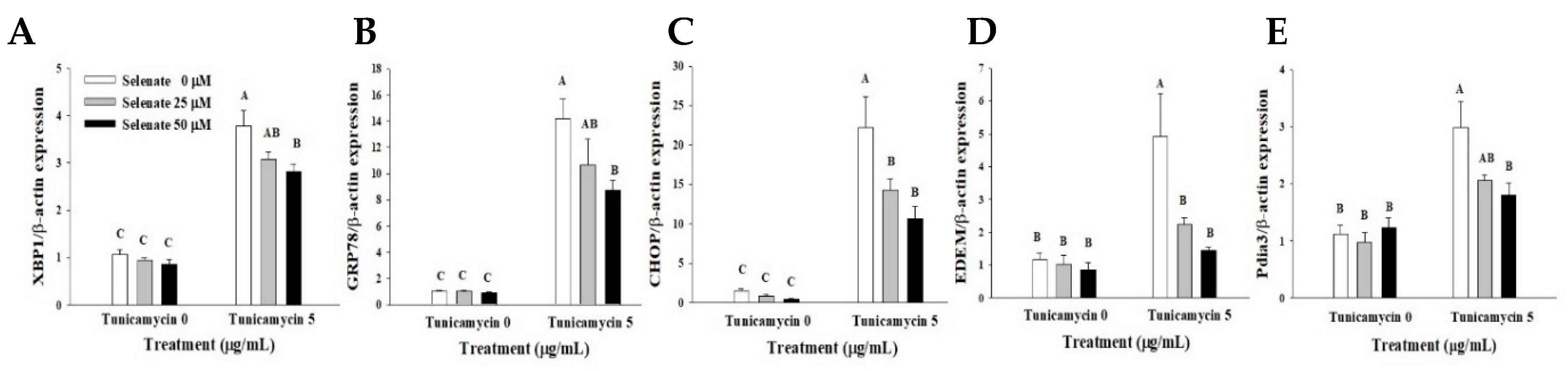
2.4. Selenate Pretreatment Inhibits ER Stress Marker Gene Expression
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Cell Culture and Differentiations of 3T3-L1 Preadipocytes
4.3. Isolation of Total RNA and Quantitative RT-PCR
4.4. Immunoblot Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Ye, R.; Jung, D.Y.; Jun, J.Y.; Li, J.; Luo, S.; Ko, H.J.; Kim, J.K.; Lee, A.S. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes 2010, 59, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, D.T.; Kaufman, R.J. A trip to the ER: Coping with stress. Trends Cell. Biol. 2004, 14, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Kozutsumi, Y.; Segal, M.; Normington, K.; Gething, M.J.; Sambrook, J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 1988, 332, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, U.; Cao, Q.; Yilmaz, E.; Lee, A.H.; Iwakoshi, N.N.; Ozdelen, E.; Tuncman, G.; Gorgun, C.; Glimcher, L.H.; Hotamisligil, G.S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 2004, 306, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Das, S.K.; Mondal, A.K.; Hackney, O.G.; Chu, W.S.; Kern, P.A.; Rasouli, N.; Spencer, H.J.; Yao-Borengasser, A.; Elbein, S.C. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. J. Clin. Endocrinol. Metab. 2008, 93, 4532–4541. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; He, Y.; Chen, H.; Wang, C.; Zenno, A.; Shi, H.; Yang, X.; Zhang, X.; Qi, L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell. Metab. 2009, 9, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Scapa, E.F.; Cohen, D.E.; Glimcher, L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008, 320, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Gregor, M.F.; Yang, L.; Fabbrini, E.; Mohammed, B.S.; Eagon, J.C.; Hotamisligil, G.S.; Klein, S. Endoplasmic reticulum stress is reduced in tissues of obese subjects after weight loss. Diabetes 2009, 58, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Basseri, S.; Lhotak, S.; Sharma, A.M.; Austin, R.C. The chemical chaperone 4-phenylbutyrate inhibits adipogenesis by modulating the unfolded protein response. J. Lipid. Res. 2009, 50, 2486–2501. [Google Scholar] [PubMed]
- Ozcan, U.; Yilmaz, E.; Ozcan, L.; Furuhashi, M.; Vaillancourt, E.; Smith, R.O.; Gorgun, C.Z.; Hotamisligil, G.S. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 2006, 313, 1137–1140. [Google Scholar] [PubMed]
- Navarro-Alarcon, M.; Cabrera-Vique, C. Selenium in food and the human body: A review. Sci. Total Environ. 2008, 400, 115–141. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.Y.; Kim, S.J.; Kim, H.G.; Lee, J.H.; Choi, Y.; Lee, H.; Kim, Y. Evaluation of estrogenicity of major heavy metals. Sci. Total Environ. 2003, 312, 15–21. [Google Scholar] [CrossRef]
- Chen, T.; Wong, Y.S. Selenocystine induces reactive oxygen species-mediated apoptosis in human cancer cells. Biomed. Pharmacother. 2009, 63, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Yang, T.B.; Wei, J.; Lei, G.; Zeng, C. Association between serum selenium level and type 2 diabetes mellitus: A non-linear dose–response meta-analysis of observational studies. Nutr. J. 2016, 15, 48. [Google Scholar] [CrossRef] [PubMed]
- Bock, B.C.; Kanarek, R.B.; Aprille, J.R. Mineral content of the diet alters sucrose-induced obesity in rats. Physiol. Behav. 1995, 57, 659–668. [Google Scholar] [CrossRef]
- Kim, C.Y.; Zhu, Y.; Buhman, K.K.; Kim, K.H. Dietary selenate attenuates adiposity and improves insulin sensitivity in high-fat diet-induced obese mice. J. Func. Foods 2015, 17, 33–42. [Google Scholar] [CrossRef]
- Kim, C.Y.; Kim, G.-N.; Wiacek, J.L.; Chen, C.Y.; Kim, K.H. Selenate inhibits adipogenesis through induction of transforming growth factor-β1 (TGF-β1) signaling. Biochem. Biophys. Res. Commun. 2012, 426, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Papp, L.V.; Lu, J.; Holmgren, A.; Khanna, K.K. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid. Redox Signal. 2007, 9, 775–806. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Walder, K.; Sunderland, T.; Kantham, L.; Feng, H.C.; Quick, M.; Bishara, N.; de Silva, A.; Augert, G.; Tenne-Brown, J.; et al. Elevation in Tanis expression alters glucose metabolism and insulin sensitivity in H4IIE cells. Diabetes 2003, 52, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Shibata, Y.; Yun, C.; Ron, D.; Rapoport, T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 2004, 429, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Shibata, Y.; Kikkert, M.; van Voorden, S.; Wiertz, E.; Rapoport, T.A. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 2005, 102, 14132–14138. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Gao, Y.; Walder, K.; Collier, G.R.; Skelton, J.; Kissebah, A.H. SEPS1 protects RAW264.7 cells from pharmacological ER stress agent-induced apoptosis. Biochem. Biophys. Res. Commun. 2007, 354, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Feng, H.C.; Walder, K.; Bolton, K.; Sunderland, T.; Bishara, N.; Quick, M.; Kantham, L.; Collier, G.R. Regulation of the selenoprotein SelS by glucose deprivation and endoplasmic reticulum stress—SelS is a novel glucose-regulated protein. FEBS Lett. 2004, 563, 185–190. [Google Scholar] [CrossRef]
- Walder, K.; Kantham, L.; McMillan, J.S.; Trevaskis, J.; Kerr, L.; De Silva, A.; Sunderland, T.; Godde, N.; Gao, Y.; Bishara, N.; et al. Tanis: A link between type 2 diabetes and inflammation? Diabetes 2002, 51, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jang, J.K.; Ko, K.Y.; Jin, Y.; Ham, M.; Kang, H.; Kim, I.Y. Degradation of selenoprotein S and selenoprotein K through PPARγ-mediated ubiquitination is required for adipocyte differentiation. Cell Death Differ. 2018, 1. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.Y.; Kim, K.H. Dexamethasone-induced selenoprotein s (SEPS1) degradation is required for adipogenesis. J. Lipid Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Graves, R.A.; Budavari, A.I.; Erdjument-Bromage, H.; Lui, M.; Hu, E.; Tempst, P.; Spiegelman, B.M. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR7 and RXRα. Nucleic Acids Res. 1994, 22, 5628–5634. [Google Scholar] [CrossRef] [PubMed]
- Rayalam, S.; Della-Fera, M.A.; Baile, C.A. Phytochemicals and regulation of the adipocyte life cycle. J. Nutr. Biochem. 2008, 19, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell. Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Sul, H.S.; Smas, C.M.; Wang, D.; Chen, L. Regulation of fat synthesis and adipose differentiation. Prog. Nucleic Acid. Res. Mol. Biol. 1998, 60, 317–345. [Google Scholar] [PubMed]
- Zhang, Y.; Chen, X. Reducing selenoprotein P expression suppresses adipocyte differentiation as a result of increased preadipocyte inflammation. Am. J. Physiol. 2011, 300, E77–E85. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.Y.; Kim, K.-H. Selenate Prevents Adipogenesis through Induction of Selenoprotein S and Attenuation of Endoplasmic Reticulum Stress. Molecules 2018, 23, 2882. https://doi.org/10.3390/molecules23112882
Kim CY, Kim K-H. Selenate Prevents Adipogenesis through Induction of Selenoprotein S and Attenuation of Endoplasmic Reticulum Stress. Molecules. 2018; 23(11):2882. https://doi.org/10.3390/molecules23112882
Chicago/Turabian StyleKim, Choon Young, and Kee-Hong Kim. 2018. "Selenate Prevents Adipogenesis through Induction of Selenoprotein S and Attenuation of Endoplasmic Reticulum Stress" Molecules 23, no. 11: 2882. https://doi.org/10.3390/molecules23112882
APA StyleKim, C. Y., & Kim, K.-H. (2018). Selenate Prevents Adipogenesis through Induction of Selenoprotein S and Attenuation of Endoplasmic Reticulum Stress. Molecules, 23(11), 2882. https://doi.org/10.3390/molecules23112882





