Comparative Transcriptomics Analysis for Gene Mining and Identification of a Cinnamyl Alcohol Dehydrogenase Involved in Methyleugenol Biosynthesis from Asarum sieboldii Miq.
Abstract
:1. Introduction
2. Results
2.1. De Novo Assembly and Sequence Annotation
2.2. Gene Ontology Classification, COG Classification, and Metabolic Pathway Assignment by KEGG
2.3. Gene Expression Analysis of AsCADs by qRT-PCR
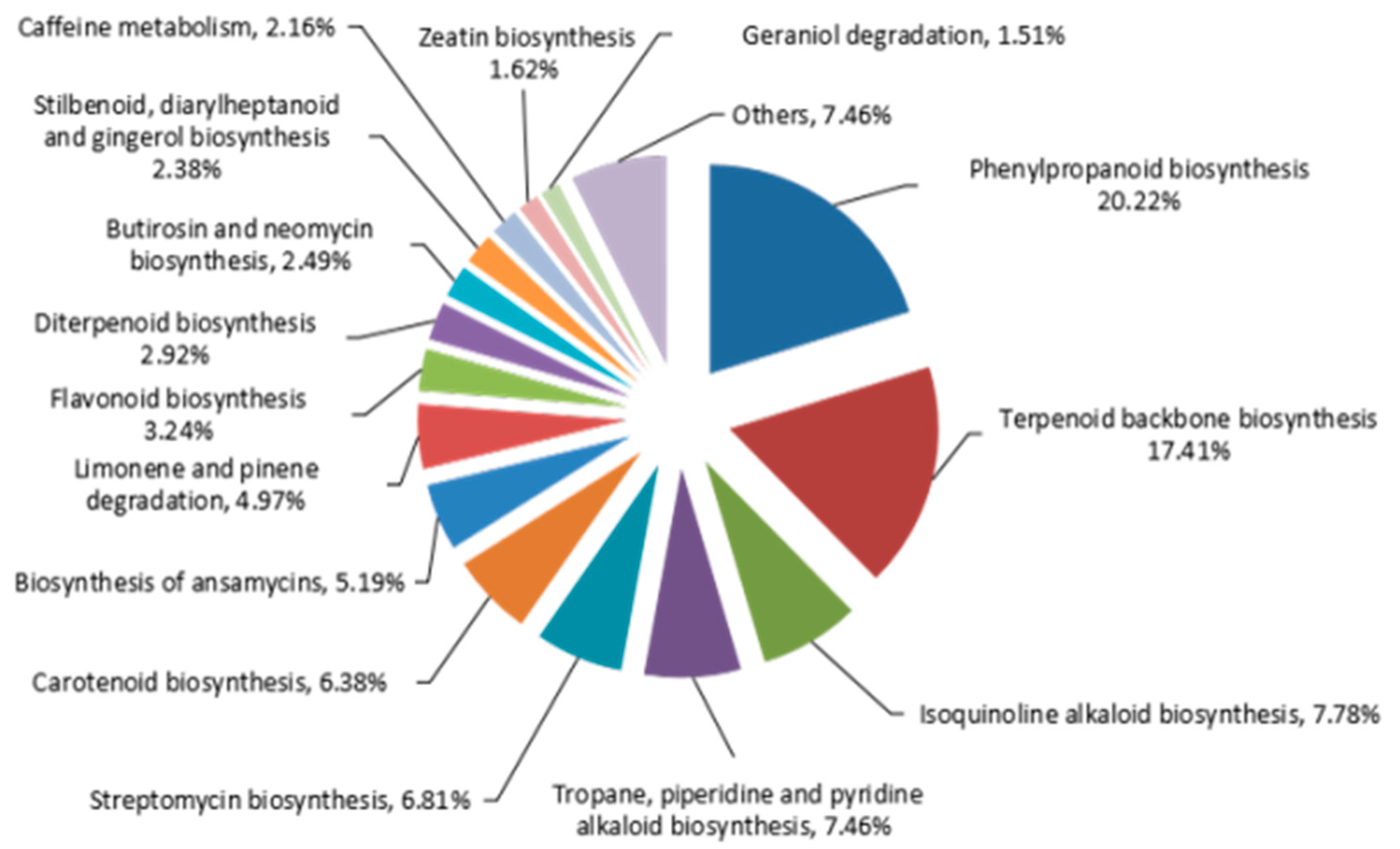
2.4. Identifying Genes Involved in Biosynthesis of Phenylpropanoids Pathway in A. sieboldii
2.5. Cloning and Phylogenetic Analysis of AsCADs
2.6. Cloning and Heterologous Expression of AsCADs
2.7. CAD Activity and Kinetic Parameters of the Recombinant AsCADs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Illumina Sequencing for Transcriptome Analysis
4.3. De Novo Assembly and Functional Annotation Analysis
4.4. Quantitative Real-Time PCR Validation
4.5. Identifying Genes Related to Biosynthesis of Methyleugenol and Asarinin in A. sieboldii
4.6. Multiple Sequence Alignments and Phylogenetic Analysis of AsCADs
4.7. Cloning of AsCADs
4.8. Heterogeneous Expression of Recombinant AsCADs
4.9. CAD Activity Assay and Determination of Kinetic Parameters
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lawrence, M.K. Phylogenetic relationships in Asarum (Aristolochiaceae) based on morphology and ITS sequences. Am. J. Bot. 1998, 85, 1454–1467. [Google Scholar]
- Michl, J.; Bello, O.; Kite, G.C.; Simmonds, M.S.J.; Heinrich, M. Medicinally used Asarum species: High-resolution LC-MS analysis of aristolochic acid analogs and in vitro toxicity screening in HK-2 cells. Front. Pharmacol. 2017, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopeia Commission. Chinese Pharmacopoeia Part I, 2015 ed.; Chemical Industry Press: Beijing, China, 2015; pp. 230–231. [Google Scholar]
- Kopyt’ko, Y.F.; Shchurevich, N.N.; Sokol’skaya, T.A.; Markaryan, A.A.; Dargaeva, T.D. Uses, chemical composition, and standardization of plant raw material and medicinal substances from plants of the genus Asarum L. Pharm. Chem. J. 2013, 47, 157–168. [Google Scholar] [CrossRef]
- Li, C.; Xu, F.; Cao, C.; Shang, M.-Y.; Zhang, C.-Y.; Yu, J.; Liu, G.-X.; Wang, X.; Cai, S.-Q. Comparative analysis of two species of Asari Radix et Rhizoma by electronic nose, headspace GC-MS and chemometrics. J. Pharm. Biomed. Anal. 2013, 85, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, J.; Zhang, F.; Li, L.; Liu, Z.; He, Z. Essential oil components from Asarum sieboldii Miquel are toxic to the house dust mite Dermatophagoides farinae. Parasitol. Res. 2012, 111, 1895–1899. [Google Scholar] [CrossRef]
- Han, Y.; Kim, S.J. Memory enhancing actions of Asiasari radix extracts via activation of insulin receptor and extracellular signal regulated kinase (ERK) I/II in rat hippocampus. Brain Res. 2003, 974, 193–201. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, J.H.; Shin, H.K.; Choi, Y.H.; Lee, J.D.; Choi, B.T. Partially purified Asiasari radix inhibits melanogenesis through extracellular signal-regulated kinase signaling in B16F10 cells. Int. J. Mol. Med. 2010, 25, 287–292. [Google Scholar] [CrossRef]
- Jeong, S.H.; Lee, J.E.; Jin, S.H.; Ko, Y.; Park, J.B. Effect of Asiasari radix on osteoblastic differentiation of stem cells derived from gingiva. J. Tradit. Chin. Med. 2016, 36, 756–759. [Google Scholar] [CrossRef]
- Kim, H.M.; Moon, Y.S. Asiasari radix inhibits immunoglobulin E production on experimental models in vitro and in vivo. Immunopharmacol. Immunotoxicol. 1999, 21, 469–481. [Google Scholar] [CrossRef]
- Lee, J.A.; Lee, M.Y.; Seo, C.S.; Ha, H.; Lee, H.; Kim, J.H. Asiasari sieboldii suppresses inflammatory mediators through the induction of hemeoxygenase-1 expression in RAW264.7 cells. Immunopharmacol. Immunotoxicol. 2012, 34, 15–20. [Google Scholar] [CrossRef]
- Oh, S.M.; Kim, J.; Lee, J.; Yi, J.M.; Oh, D.S.; Bang, O.S.; Kim, N.S. Anticancer potential of an ethanol extract of Asiasari radix against HCT-116 human colon cancer cells in vitro. Oncol. Lett. 2013, 5, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Nakai, M.; Fukui, Y.; Tomimori, N.; Fukuchi-Mizutani, M.; Saito, M.; Satake, H.; Tanaka, T.; Katsuta, M.; Umezawa, T.; et al. Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. Proc. Natl. Acad. Sci. USA 2006, 103, 10116–10121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathak, N.; Bhaduri, A.; Bhat, K.V.; Rai, A.K. Tracking sesamin synthase gene expression through seed maturity in wild and cultivated sesame species—A domestication footprint. BMC Plant Biol. 2015, 17, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Bedgar, D.L.; Katayama, T.; Lewis, N.G. On the stereoselective synthesis of (+)-pinoresinol in Forsythia suspensa from its achiral precursor, coniferyl alcohol. Phytochemistry 1992, 31, 3869–3874. [Google Scholar] [CrossRef] [PubMed]
- Halls, S.C.; Davin, L.B.; Kramer, D.M.; Lewis, N.G. Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry 2004, 43, 2587–2595. [Google Scholar] [CrossRef] [PubMed]
- Kazenwadel, C.; Klebensberger, J.; Richter, S.; Pfannstiel, J.; Gerken, U.; Pickel, B.; Schaller, A.; Hauer, B. Optimized expression of the dirigent protein AtDIR6 in Pichia pastoris and impact of glycosylation on protein structure and function. Appl. Microbiol. Biotechnol. 2013, 97, 7215–7227. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W.; et al. A novel Soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef] [PubMed]
- Pickel, B.; Pfannstiel, J.; Steudle, A.; Lehmann, A.; Gerken, U.; Pleiss, J.; Schaller, A. A model of dirigent proteins derived from structural and functional similarities with allene oxide cyclase and lipocalins. FEBS J. 2012, 279, 1980–1993. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.S.; Hattori, M. A new mammalian lignan precursor, asarinin. Food Chem. 2011, 124, 895–899. [Google Scholar] [CrossRef]
- Kushiro, M.; Masaoka, T.; Hageshita, S.; Takahashi, Y.; Ide, T.; Sugano, M. Comparative effect of sesamin and episesamin on the activity and gene expression of enzymes in fatty acid oxidation and synthesis in rat liver. J. Nutr. Biochem. 2002, 13, 289–295. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, L.; Wang, Z.; Chen, Y.; Zhang, G.; Zhang, D.; Wang, X.; Bai, X.; Li, X.; Lili, Z. The effect of asarinin on toll-like pathway in rats after cardiac allograft implantation. Transplant. Proc. 2015, 47, 545–548. [Google Scholar] [CrossRef]
- Jeong, M.; Kim, H.M.; Lee, J.S.; Choi, J.H.; Jang, D.S. (−)-Asarinin from the roots of Asarum sieboldii induces apoptotic cell death via caspase activation in human ovarian cancer cells. Molecules 2018, 23, 1849. [Google Scholar] [CrossRef]
- Kosuge, T.; Yokota, M.; Nagasawa, M.; Nukaya, H.; Gotoh, Y. Studies on antitussive principles of Asiasari Radix. Chem. Pharm. Bull. 1978, 26, 2284–2285. [Google Scholar] [CrossRef]
- Yano, S.; Suzuki, Y.; Yuzurihara, M.; Kase, Y.; Takeda, S.; Watanabe, S.; Aburada, M.; Miyamoto, K. Antinociceptive effect of methyleugenol on formalin-induced hyperalgesia in mice. Eur. J. Pharmacol. 2006, 553, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Yan, Y.; Yan, Y.; Deng, S.; Liu, Y.M.; Fan, H.R.; Ma, B.; Meng, B.; Mei, B.; Li, W.G.; Li, F. Methyleugenol counteracts anorexigenic signals in association with GABAergic inhibition in the central amygdala. Neuropharmacology 2018, 141, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Yauk, Y.K.; Souleyre, E.J.F.; Matich, A.J.; Chen, X.; Wang, M.Y.; Plunkett, B.; Dare, A.P.; Espley, R.V.; Tomes, S.; Chagné, D.; et al. Alcohol acyl transferase 1 links two distinct volatile pathways that produce esters and phenylpropenes in apple fruit. Plant J. 2017, 91, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Jayaramaiah, R.H.; Beedkar, S.D.; Singh, P.A.; Joshi, R.S.; Mulani, F.A.; Dholakia, B.B.; Punekar, S.A.; Gade, W.N.; Thulasiram, H.V.; Giri, A.P. Comparative functional characterization of eugenol synthase from four different Ocimum species: Implications on eugenol accumulation. Biochim. Biophys. Acta 2016, 1864, 1539–1547. [Google Scholar] [CrossRef]
- Gupta, A.K.; Schauvinhold, I.; Pichersky, E.; Schiestl, F.P. Eugenol synthase genes in floral scent variation in Gymnadenia species. Funct. Integr. Genom. 2014, 14, 779–788. [Google Scholar] [CrossRef]
- Yan, H.; Baudino, S.; Caissard, J.C.; Florence, N.; Zhang, H.; Tang, K.; Li, S.; Lu, S. Functional characterization of the eugenol synthase gene (RcEGS1) in rose. Plant Physiol. Biochem. 2018, 129, 21–26. [Google Scholar] [CrossRef]
- Koeduka, T.; Kajiyama, M.; Furuta, T.; Suzuki, H.; Tsuge, T.; Matsui, K. Characterization of an O-methyltransferase specific to guaiacol-type benzenoids from the flowers of loquat (Eriobotrya japonica). J. Biosci. Bioeng. 2016, 122, 679–684. [Google Scholar] [CrossRef]
- Koeduka, T.; Fridman, E.; Gang, D.R.; Vassao, D.G.; Jackson, B.L.; Kish, C.M.; Orlova, I.; Spassova, S.M.; Lewis, N.G.; Noel, J.P.; et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 2006, 103, 10128–10133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renu, I.K.; Haque, I.; Kumar, M.; Poddar, R.; Bandopadhyay, R.; Rai, A.; Mukhopadhyay, K. Characterization and functional analysis of eugenol O-methyltransferase gene reveal metabolite shifts, chemotype specific differential expression and developmental regulation in Ocimum tenuiflorum L. Mol. Biol. Rep. 2014, 41, 1857–1870. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pichersky, E. Characterization of S-adenosyl-l-methionine: (Iso)eugenol O-methyltransferase involved in floral scent production in Clarkia breweri. Arch. Biochem. Biophys. 1998, 349, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Yauk, Y.K.; Chagne, D.; Tomes, S.; Matich, A.J.; Wang, M.Y.; Chen, X.; Maddumage, R.; Hunt, M.B.; Rowan, D.D.; Atkinson, R.G. The O-methyltransferase gene MdoOMT1 is required for biosynthesis of methylated phenylpropenes in ripe apple fruit. Plant J. 2015, 82, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Boudet, A.M.; Lapierre, C.; Grima-Pettenati, J. Biochemistry and molecular biology of lignification. New Phytol. 1995, 129, 203–236. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.J.; Kim, M.R.; Bedgar, D.L.; Moinuddin, S.G.; Cardenas, C.L.; Davin, L.B.; Kang, C.; Lewis, N.G. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Dorairaj, D.; Ismail, M.R. Distribution of silicified microstructures, regulation of cinnamyl alcohol dehydrogenase and lodging resistance in silicon and paclobutrazol mediated Oryza sativa. Front. Physiol. 2017, 8, 491. [Google Scholar] [CrossRef]
- Geissler, M.; Burghard, M.; Volk, J.; Staniek, A.; Warzecha, H. A novel cinnamyl alcohol dehydrogenase (CAD)-like reductase contributes to the structural diversity of monoterpenoid indole alkaloids in Rauvolfia. Planta 2016, 243, 813–824. [Google Scholar] [CrossRef]
- Ozparpucu, M.; Gierlinger, N.; Burgert, I.; Van Acker, R.; Vanholme, R.; Boerjan, W.; Pilate, G.; Déjardin, A.; Rüggeberg, M. The effect of altered lignin composition on mechanical properties of cinnamyl alcohol dehydrogenase (CAD) deficient poplars. Planta 2018, 247, 887–897. [Google Scholar] [CrossRef]
- Ikezawa, N.; Iwasa, K.; Sato, F. CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 2009, 28, 123–133. [Google Scholar] [CrossRef]
- Saathoff, A.J.; Hargrove, M.S.; Haas, E.J.; Tobias, C.M.; Twigg, P.; Sattler, S.; Sarath, G. Switchgrass PviCAD1: Understanding residues important for substrate preferences and activity. Appl. Biochem. Biotechnol. 2012, 168, 1086–1100. [Google Scholar] [CrossRef]
- Barakat, A.; Bagniewska-Zadworna, A.; Choi, A.; Plakkat, U.; DiLoreto, D.S.; Yellanki, P.; Carlson, J.E. The cinnamyl alcohol dehydrogenase gene family in Populus: Phylogeny, organization, and expression. BMC Plant Biol. 2009, 9, 26:1–26:15. [Google Scholar] [CrossRef]
- Guo, D.M.; Ran, J.H.; Wang, X.Q. Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: The emergence of real lignin is associated with the origin of bona fide CAD. J. Mol. Evol. 2010, 71, 202–218. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, C.; Liu, W.; Qi, H.; Chen, H.; Cao, S. The cinnamyl alcohol dehydrogenase gene family in melon (Cucumis melo L.): Bioinformatic analysis and expression patterns. PLoS ONE 2014, 9, 101730. [Google Scholar] [CrossRef]
- Rong, W.; Luo, M.; Shan, T.; Wei, X.; Du, L.; Xu, H.; Zhang, Z. A wheat cinnamyl alcohol dehydrogenase TaCAD12 contributes to host resistance to the sharp eyespot disease. Front. Plant Sci. 2016, 7, 1723. [Google Scholar] [CrossRef]
- Saballos, A.; Ejeta, G.; Sanchez, E.; Kang, C.; Vermerris, W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the Brown midrib6 gene. Genetics 2009, 181, 783–795. [Google Scholar] [CrossRef]
- Marques, J.V.; Kim, K.W.; Lee, C.; Costa, M.A.; May, G.D.; Crow, J.A.; Davin, L.B.; Lewis, N.G. Next generation sequencing in predicting gene function in podophyllotoxin biosynthesis. J. Biol. Chem. 2013, 288, 466–479. [Google Scholar] [CrossRef]
- Ikezawa, N.; Tanaka, M.; Nagayoshi, M.; Shinkyo, R.; Sakaki, T.; Inouye, K.; Sato, F. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 2003, 278, 38557–38565. [Google Scholar] [CrossRef]
- Diaz Chavez, M.L.; Rolf, M.; Gesell, A.; Kutchan, T.M. Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 2011, 507, 186–193. [Google Scholar] [CrossRef]
- Li, C.Y.; Chow, T.J.; Wu, T.S. The epimerization of sesamin and asarinin. J. Nat. Prod. 2005, 68, 1622–1624. [Google Scholar] [CrossRef]
- Bos, R.; Hendriks, H.; van Os, F.H. The composition of the essential oil in the leaves of Coleus aromaticus Bentham and their importance as a component of the species antiaphthosae Ph. Ned. Ed. V. Pharm. Weekbl. Sci. 1983, 5, 129–130. [Google Scholar] [CrossRef]
- Chokechaijaroenporn, O.; Bunyapraphatsara, N.; Kongchuensin, S. Mosquito repellent activities of Ocimum volatile oils. Phytomedicine 1994, 1, 135–139. [Google Scholar] [CrossRef]
- Da Silva, J.K.; Andrade, E.H.; Guimaraes, E.F.; Maia, J.G. Essential oil composition, antioxidant capacity and antifungal activity of Piper divaricatum. Nat. Prod. Commun. 2010, 5, 477–480. [Google Scholar]
- De Vincenzi, M.; Silano, M.; Stacchini, P.; Scazzocchio, B. Constituents of aromatic plants: I. Methyleugenol. Fitoterapia 2000, 71, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Yanagisawa, T.; Okui, Y.; Ikeya, Y.; Maruno, M.; Fujita, T. Studies on anti-allergic components in the roots of Asiasarum sieboldi. Planta Med. 1994, 60, 124–127. [Google Scholar] [CrossRef]
- Kamdem, D.P.; Gage, D.A. Chemical composition of essential oil from the root bark of Sassafras albidum. Planta Med. 1995, 61, 574–575. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, M.; Luo, W.; Yang, B.; Jiang, Y. Identification of volatile components in Phyllanthus emblica L. and their antimicrobial activity. J. Med. Food 2009, 12, 423–428. [Google Scholar] [CrossRef]
- Medina, A.L.; Lucero, M.E.; Holguin, F.O.; Estell, R.E.; Posakony, J.J.; Simon, J.; O’Connell, M.A. Composition and antimicrobial activity of Anemopsis californica leaf oil. J. Agric. Food Chem. 2005, 53, 8694–8698. [Google Scholar] [CrossRef]
- Miele, M.; Ledda, B.; Falugi, C.; Mazzei, M. Methyleugenol and eugenol variation in Ocimum basilicum cv. Genovese gigante grown in greenhouse and in vitro. Boll. Soc. Ital. Biol. Sper. 2001, 77, 43–50. [Google Scholar]
- Pino Benitez, N.; Melendez Leon, E.M.; Stashenko, E.E. Eugenol and methyl eugenol chemotypes of essential oil of species Ocimum gratissimum L. and Ocimum campechianum Mill. from Colombia. J. Chromatogr. Sci. 2009, 47, 800–803. [Google Scholar] [CrossRef] [PubMed]
- Sayyah, M.; Valizadeh, J.; Kamalinejad, M. Anticonvulsant activity of the leaf essential oil of Laurus nobilis against pentylenetetrazole- and maximal electroshock-induced seizures. Phytomedicine 2002, 9, 212–216. [Google Scholar] [CrossRef]
- Schiestl, R.H.; Chan, W.S.; Gietz, R.D.; Mehta, R.D.; Hastings, P.J. Safrole, eugenol and methyleugenol induce intrachromosomal recombination in yeast. Mutat. Res. 1989, 224, 427–436. [Google Scholar] [CrossRef]
- Chen, C.; Spriano, D.; Lehmann, T.; Meier, B. Reduction of safrole and methyleugenol in Asari radix et rhizoma by decoction. Forsch. Komplementmed. 2009, 16, 162–166. [Google Scholar] [CrossRef]
- Li, X.; Ma, D.; Chen, J.; Pu, G.; Ji, Y.; Lei, C.; Du, Z.; Liu, B.; Ye, H.; Wang, H. Biochemical characterization and identification of a cinnamyl alcohol dehydrogenase from Artemisia annua. Plant Sci. 2012, 193–194, 85–95. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Lethiec, F.; Duroux, P.; Gascuel, O. PhyML Online—A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Saathoff, A.J.; Sarath, G.; Chow, E.K.; Dien, B.S.; Tobias, C.M. Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 2011, 6, 16416. [Google Scholar] [CrossRef]
Sample Availability: Not available. |







| Public Database | Number of Unigenes Annotated in Asarum sieboldii |
|---|---|
| Nr | 127,167 |
| Swiss-Prot | 76,685 |
| COG | 22,386 |
| KEGG | 35,326 |
| Enzyme Kinetic Parameters | Coniferyl Aldehyde | Sinapyl Aldehyde | P-coumaryl Aldehyde |
|---|---|---|---|
| Km (µM) | 27.32 ± 6.186 | 35.02 ± 8.043 | 45.52 ± 8.888 |
| Vmax (nmol s−1 mg protein−1) | 342 ± 26.29 | 448.5 ± 50.54 | 383.8 ± 31.63 |
| Kcat (s−1) | 221.7 | 290.8 | 248.8 |
| Kcat/Km (µM−1 s−1) | 8.115 | 8.304 | 5.466 |
| ▲ 48 | ● 75 | ● 86 | ▲ 94 | ■ 118 | ■ 122 | ● 275 | ● 276 | ● 285 | ● 299 | ● 300 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AaCAD | T | V | V | C | W | D | V | I | P | F | I |
| AtCAD5 | T | V | V | C | W | D | V | I | P | F | I |
| AsCAD5 | A | Q | I | T | 119T | 123S | 276F | 277P | G | V | T |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Xu, C.; Zhang, H.; Liu, F.; Ma, D.; Liu, Z. Comparative Transcriptomics Analysis for Gene Mining and Identification of a Cinnamyl Alcohol Dehydrogenase Involved in Methyleugenol Biosynthesis from Asarum sieboldii Miq. Molecules 2018, 23, 3184. https://doi.org/10.3390/molecules23123184
Liu J, Xu C, Zhang H, Liu F, Ma D, Liu Z. Comparative Transcriptomics Analysis for Gene Mining and Identification of a Cinnamyl Alcohol Dehydrogenase Involved in Methyleugenol Biosynthesis from Asarum sieboldii Miq. Molecules. 2018; 23(12):3184. https://doi.org/10.3390/molecules23123184
Chicago/Turabian StyleLiu, Jinjie, Chong Xu, Honglei Zhang, Fawang Liu, Dongming Ma, and Zhong Liu. 2018. "Comparative Transcriptomics Analysis for Gene Mining and Identification of a Cinnamyl Alcohol Dehydrogenase Involved in Methyleugenol Biosynthesis from Asarum sieboldii Miq." Molecules 23, no. 12: 3184. https://doi.org/10.3390/molecules23123184





