Synthesis, Biological Evaluation and Docking Studies of Benzoxazoles Derived from Thymoquinone
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Antibacterial Activity
2.3. Antifungal Activity
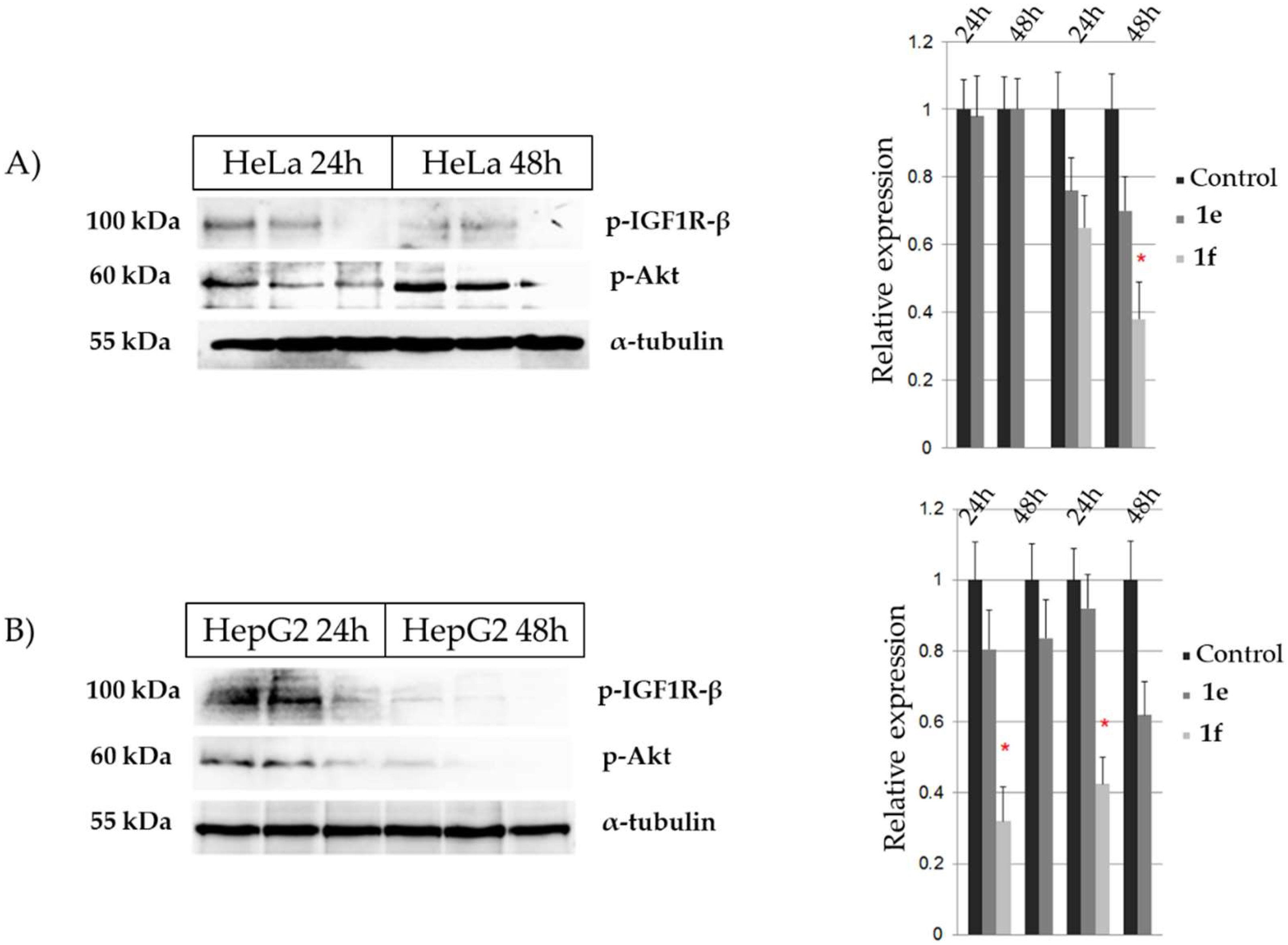
2.4. Cytotoxicity
2.5. Docking Studies
3. Materials and Methods
3.1. General Experimental Procedures
3.2. General Procedure for the Synthesis of 3-aminothymoquinone (ATQ)
General Procedure for the Synthesis of Benzoxazole Derivatives 1a–1j
3.3. Biological Assays
3.3.1. Antimicrobial Activity
Compounds and Microorganisms
Diffusion and Micro-Dilution Assays
3.3.2. Cytotoxic Activity
Compounds and Cell Lines
MTT Assay
Western Blot
3.4. Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gholamnezhad, Z.; Havakhah, S.; Boskabady, M.H. Preclinical and clinical effects of Nigella sativa and its constituent, thymoquinone: A review. J. Ethnopharmacol. 2016, 190, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Taborsky, J.; Kunt, M.; Kloucek, P.; Lachman, J.; Zeleny, V.; Kokoska, L. Identification of potential sources of thymoquinone and related compounds in Asteraceae, Cupressaceae, Lamiaceae, and Ranunculaceae families. Cent. Eur. J. Chem. 2012, 10, 1899–1906. [Google Scholar] [CrossRef]
- Pang, J.; Shen, N.; Yan, F.; Zhao, N.; Dou, L.; Wu, LC.; Seiler, CL.; Yu, L.; Yang, K.; Bachanova, V.; et al. Thymoquinone exerts potent growth-suppressive activity on leukemia through DNA hypermethylation reversal in leukemia cells. Oncotarget 2017, 8, 34453–34467. [Google Scholar] [CrossRef] [PubMed]
- El-Far, AH. Thymoquinone Anticancer Discovery: Possible Mechanisms. Curr. Drug. Discov. Technol. 2015, 12, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Attoub, S.; Sperandio, O.; Raza, H.; Arafat, K.; Al-Salam, S.; Al Sultan, M.A.; Al Safi, M.; Takahashi, T.; Adem, A. Thymoquinone as an anticancer agent: Evidence from inhibition of cancer cells viability and invasion in vitro and tumor growth in vivo. Fundam. Clin. Pharmacol. 2013, 27, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.; Chun, K.S.; Aruoma, O.I.; Kundu, J.K. Mechanistic perspectives on cancer chemoprevention/chemotherapeutic effects of thymoquinone. Mutat. Res. 2014, 768, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Stock, R.; Fakhoury, I.H.; Zaki, A.M.; El-Baba, C.O.; Gali-Muhtasib, HU. Thymoquinone: Fifty years of success in the battle against cancer models. Drug Discov. Today. 2014, 19, 18–30. [Google Scholar] [CrossRef]
- Randhawa, M.A.; Alenazy, A.K.; Alrowaili, M.G.; Basha, J. An active principle of Nigella sativa L., thymoquinone, showing significant antimicrobial activity against anaerobic bacteria. J. Intercult. Ethnopharmacol. 2016, 6, 97–101. [Google Scholar] [CrossRef]
- Chaieb, K.; Kouidhi, B.; Jrah, H.; Mahdouani, K.; Bakhrouf, A. Antibacterial activity of Thymoquinone, an active principle of Nigella sativa and its potency to prevent bacterial biofilm formation. BMC Complement. Altern. Med. 2011, 11, 29. [Google Scholar] [CrossRef]
- Goel, S.; Mishra, P. Thymoquinone inhibits biofilm formation and has selective antibacterial activity due to ROS generation. Appl. Microbiol. Biotechnol. 2018, 102, 1955–1967. [Google Scholar] [CrossRef]
- Khan, M.A.; Younus, H. Thymoquinone shows the diverse therapeutic actions by modulating multiple cell signaling pathways: Single drug for multiple targets. Curr. Pharm. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Majdalawieh, A.F.; Fayyad, M.W. Immunomodulatory and anti-inflammatory action of Nigella sativa and thymoquinone: A comprehensive review. Int. Immunopharmacol. 2015, 28, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Elmaci, I.; Altinoz, M.A. Thymoquinone: An edible redox-active quinone for the pharmacotherapy of neurodegenerative conditions and glial brain tumors. A short review. Biomed. Pharmacother. 2016, 83, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Azmi, A.S.; Padhye, S.; Singh, M.W.; Baruah, J.B.; Philip, P.A.; Sarkar, F.H.; Mohammad, R.M. Structure-activity studies on therapeutic potential of Thymoquinone analogs in pancreatic cancer. Pharm. Res. 2010, 27, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Breyer, S.; Effenberger, K.; Schobert, R. Effects of thymoquinone–fatty acid conjugates on cancer cells. Chem. Med. Chem. 2009, 4, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Wirries, A.; Breyer, S.; Quint, K.; Schobert, R.; Ocker, M. Thymoquinone hydrazone derivatives cause cell cycle arrest in p53-competent colorectal cancer cells. Exp. Ther. Med. 2010, 1, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Odeh, F.; Ismail, S.I.; Abu-Dahab, R.; Mahmoud, I.S.; Al Bawab, A. Thymoquinone in liposomes: A study of loading efficiency and biological activity towards breast cancer. Drug Deliv. 2012, 19, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Ganea, G.M.; Fakayode, S.O.; Losso, J.N.; van Nostrum, C.F.; Sabliov, C.M.; Warner, I.M. Delivery of phytochemical thymoquinone using molecular micelle modified poly (D, L lactide-co-glycolide)(PLGA) nanoparticles. Nanotechnology 2010, 21, 285104. [Google Scholar] [CrossRef] [PubMed]
- Demmer, C.S.; Bunch, L. Benzoxazoles and oxazolopyridines in medicinal chemistry studies. Eur. J. Med. Chem. 2015, 97, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Manjunath, B.; Ghorai, S.; Sasmal, S. Benzoxazole Alkaloids: Occurrence, Chemistry, and Biology. Alkaloids Chem. Biol. 2018, 79, 71–137. [Google Scholar]
- Oksuzoglu, E.; Tekiner-Gulbas, B.; Alper, S.; Temiz-Arpaci, O.; Ertan, T.; Yildiz, I.; Diril, N.; Sener-Aki, E.; Yalcin, I. Some benzoxazoles and benzimidazoles as DNA topoisomerase I and II inhibitors. J. Enzyme Inhib. Med. Chem. 2008, 23, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Jacob, M.R.; Reynolds, M.B.; Kerwin, S.M. Synthesis and evaluation of anticancer benzoxazoles and benzimidazoles related to UK-1. Bioorg. Med. Chem. 2002, 10, 3997–4004. [Google Scholar] [CrossRef]
- Wang, B.B.; Maghami, N.; Goodlin, V.L.; Smith, P.J. Critical structural motif for the catalytic inhibition of human topoisomerase II by UK-1 and analogs. Bioorg. Med. Chem. Lett. 2004, 14, 3221–3226. [Google Scholar] [CrossRef] [PubMed]
- Rida, S.M.; Ashour, F.A.; El-Hawash, S.A.M.; ElSemary, M.M.; Badr, M.H.; Shalaby, M.A. Synthesis of some novel benzoxazole derivatives as anticancer, anti-HIV-1 and antimicrobial agents. Eur. J. Med. Chem. 2005, 40, 949–959. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Lee, E.; Yu, Y.; Yun, J.; Lee, M.Y.; Kang, J.S.; Kim, W.Y.; Jeon, R. Design and synthesis of novel benzoxazole analogs as Aurora B kinase inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Paramashivappa, R.; Kumar, P.P.; Rao, P.S.; Rao, A.S. Design, synthesis and biological evaluation of benzimidazole/benzothiazole and benzoxazole derivatives as cyclooxygenase inhibitors. Bioorg. Med. Chem. Lett. 2003, 13, 657–660. [Google Scholar] [CrossRef]
- Hall, I.H.; Peaty, N.J.; Henry, J.R.; Easmon, J.; Heinisch, G.; Pürstinger, G. Investigations on the Mechanism of Action of the Novel Antitumor Agents 2-Benzothiazolyl, 2-Benzoxazolyl, and 2-Benzimidazolyl Hydrazones Derived from 2-Acetylpyridine. Arch. Pharm. (Weinheim) 1999, 332, 115–123. [Google Scholar] [CrossRef]
- Sheng, C.; Che, X.; Wang, W.; Wang, S.; Cao, Y.; Yao, J.; Miao, Z.; Zhang, W. Design and synthesis of antifungal benzoheterocyclic derivatives by scaffold hopping. Eur. J. Med. Chem. 2011, 46, 1706–1712. [Google Scholar] [CrossRef]
- Bray, H.G.; Clowes, R.C.; Thorpe, W.V. The metabolism of aminophenols, o-formamidophenol, benzoxazole, 2-methyl-and 2-phenyl-benzoxazoles and benzoxazolone in the rabbit. Biochem. J. 1952, 51, 70. [Google Scholar] [CrossRef] [PubMed]
- McElhinny, C.J.; Lewin, A.H.; Mascarella, S.W.; Runyon, S.; Brieaddy, L.; Carroll, F.I. Hydrolytic instability of the important orexin 1 receptor antagonist SB-334867: Possible confounding effects on in vivo and in vitro studies. Bioorg. Med. Chem. Lett. 2012, 22, 6661–6664. [Google Scholar] [CrossRef]
- Yusufi, M.; Banerjee, S.; Mohammad, M.; Khatal, S.; Venkateswara Swamy, K.; Khan, E.M.; Aboukameel, A.; Sarkar, F.H.; Padhye, S. Synthesis, characterization and anti-tumor activity of novel thymoquinone analogs against pancreatic cancer. Bioorg. Med. Chem. Lett. 2013, 23, 3101–3104. [Google Scholar] [CrossRef] [PubMed]
- Slater, J.W.; Steel, P.J. Syntheses of new binucleating heterocyclic ligands. Tetrahedron Lett. 2006, 47, 6941–6943. [Google Scholar] [CrossRef]
- Lee, H.; Theodorakis, E. Houben-Weyl Methods in Molecular Transformations. In Science of Synthesis; Griesbeck, A.G., Ed.; Georg Thieme: Stuttgart, Germany, 2006; Volume 28, pp. 71–86. [Google Scholar]
- Strutt, N.L.; Zhang, H.; Schneebeli, S.T.; Stoddart, J.F. Amino-Functionalized Pillar [5] arene. Chem. Eur. J. 2014, 20, 10996–11004. [Google Scholar] [CrossRef] [PubMed]
- Van Aeken, S.; Deblander, J.; De Houwer, J.; Mosselmans, T.; Abbaspour Tehrani, K. Unexpected reaction of 2-amino-1,4-naphthoquinone with aldehydes: New synthesis of naphtho[2,1-d]oxazole compounds. Tetrahedron 2011, 67, 512–517. [Google Scholar] [CrossRef]
- Li, X.; Bian, J.; Wang, N.; Qian, X.; Gu, J.; Mu, T.; Fan, J.; Yang, X.; Li, S.; Yang, T.; et al. Novel naphtho [2, 1-d] oxazole-4, 5-diones as NQO1 substrates with improved aqueous solubility: Design, synthesis, and in vivo antitumor evaluation. Bioorg. Med. Chem. 2016, 24, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Weider, P.R.; Hegedus, L.S.; Asada, H. Oxidative cyclization of unsaturated aminoquinones. Synthesis of quinolinoquinones. Palladium-catalyzed synthesis of pyrroloindoloquinones. J. Org. Chem. 1985, 50, 4276–4281. [Google Scholar] [CrossRef]
- Boojar, M.M.A.; Goodarzi, F. Cytotoxicity and the levels of oxidative stress parameters in WI38 cells following 2 macrocyclic crown ethers treatment. Clin. Chim. Acta 2006, 364, 321–327. [Google Scholar] [CrossRef]
- Arafa, E.S.A.; Zhu, Q.; Shah, Z.I.; Wani, G.; Barakat, B.M.; Racoma, I.; El-Mahdy, M.A.; Wani, A.A. Thymoquinone up-regulates PTEN expression and induces apoptosis in doxorubicin-resistant human breast cancer cells. Mutat. Res. 2011, 706, 28–35. [Google Scholar] [CrossRef]
- Hussain, A.R.; Ahmed, M.; Ahmed, S.; Manogaran, P.; Platanias, L.C.; Alvi, S.N.; Al-Kuraya, K.S.; Uddin, S. Thymoquinone suppresses growth and induces apoptosis via generation of reactive oxygen species in primary effusion lymphoma. Free Radic. Biol. Med. 2011, 50, 978–987. [Google Scholar] [CrossRef]
- Rajput, S.; Kumar, B.N.P.; Sarkar, S.; Das, S.; Azab, B.; Santhekadur, P.K.; Das, S.K.; Emdad, L.; Sarkar, D.; Fisher, P.B.; et al. Targeted apoptotic effects of thymoquinone and tamoxifen on XIAP mediated Akt regulation in breast cancer. PLoS ONE 2013, 8, e61342. [Google Scholar] [CrossRef]
- Yi, T.; Cho, S.-G.; Yi, Z.; Pang, X.; Rodriguez, M.; Wang, Y.; Sethi, G.; Aggarwal, B.B.; Liu, M. Thymoquinone inhibits tumor angiogenesis and tumor growth through suppressing AKT and extracellular signal-regulated kinase signaling pathways. Mol. Cancer Ther. 2008, 7, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Nithya, G.; Sakthisekaran, D. In silico docking studies on the anti-cancer effect of thymoquinone on interaction with phosphatase and tensin homolog located on chromosome 10q23: A regulator of PI3K/AKT pathway. Asian J. Pharm. Clin. Res. 2015, 8, 192–195. [Google Scholar]
- Brahmkhatri, V.P.; Prasanna, C.; Atreya, H.S. Insulin-like growth factor system in cancer: Novel targeted therapies. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Pourpak, A.; Morris, S.W. Inhibition of the insulin-like growth factor-1 receptor (IGF1R) tyrosine kinase as a novel cancer therapy approach. J. Med. Chem. 2009, 52, 4981–5004. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Bramley, R.L.; Cowell, I.G.; Jackson, G.H.; Austin, C.A. Proteasomal inhibition potentiates drugs targeting DNA topoisomerase II. Biochem. Pharmacol. 2016, 103, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, J.; Meng, Q.; Niu, G. Multidrug resistance protein and topoisomerase 2 alpha expression in non-small cell lung cancer are related with brain metastasis postoperatively. Int. J. Clin. Exp. Pathol. 2015, 8, 11537. [Google Scholar] [PubMed]
- Ashley, R.E.; Osheroff, N. Natural products as topoisomerase II poisons: Effects of thymoquinone on DNA cleavage mediated by human topoisomerase Iiα. Chem. Res. Toxicol. 2014, 27, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Roepke, M.; Diestel, A.; Bajbouj, K.; Walluscheck, D.; Schonfeld, P.; Roessner, A.; Schneider-Stock, R.; Gali-Muhtasib, H. Lack of p53 augments thymoquinone-induced apoptosis and caspase activation in human osteosarcoma cells. Cancer Biol. Ther. 2007, 6, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Sethi, G.; Ahn, K.S.; Aggarwal, B.B. Targeting nuclear factor-κB activation pathway by thymoquinone: Role in suppression of antiapoptotic gene products and enhancement of apoptosis. Mol. Cancer Res. 2008, 6, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Altomare, A.; Cascarano, G.; Giacovazzo, C.; Guagliardi, A.; Burla, M.C.; Polidori, G.; Camalli, M. SIR92—A program for automatic solution of crystal structures by direct methods. J. Appl. Crystallogr. 1994, 27, 435. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3. [Google Scholar] [CrossRef] [PubMed]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; Streek, J.V. Mercury: Visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Gazivoda, T.; Raić-Malić, S.; Kristafor, V.; Makuc, D.; Plavec, J.; Bratulić, S.; Kraljević-Pavelić, S.; Pavelić, K.; Naesens, L.; Andrei, G.; et al. Synthesis, cytostatic and anti-HIV evaluations of the new unsaturated acyclic C-5 pyrimidine nucleoside analogues. Bioorg. Med. Chem. 2008, 16, 5624–5634. [Google Scholar] [CrossRef] [PubMed]
- Gore, S.; Sanz García, E.; Hendrickx, P.M.S.; Gutmanas, A.; Westbrook, J.D.; Yang, H.; Feng, Z.; Baskaran, K.; Berrisford, J.M.; Hudson, B.P.; et al. Validation of structures in the Protein Data Bank. Structure 2017, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide protein data bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Strawn, L.M.; McMahon, G.; App, H.; Schreck, R.; Kuchler, W.R.; Longhi, M.P.; Hui, T.H.; Tang, C.; Levitzki, A.; Gazit, A.; et al. Flk-1 as a target for tumor growth inhibition. Cancer Res. 1996, 56, 3540–3545. [Google Scholar]
Sample Availability: Samples of the compounds ATQ and 1a–1j are available from the authors. |




| Bacteria | Compound | ||
|---|---|---|---|
| TQ | ATQ | Amikacin | |
| MIC (μM) | |||
| E. coli | 29.84 | 111.59 | 8.54 |
| P. aeruginosa | 59.68 | 217.61 | 85.38 |
| P. hauseri | 29.84 | 217.61 | 11.95 |
| K. pneumoniae | 59.68 | 435.22 | 13.71 |
| S. aureus | 29.84 | 111.59 | 18.78 |
| B. subtilis | 59.68 | 217.61 | 71.72 |
| C. sporogenes | 121.80 | 435.22 | 25.61 |
| M. luteus | 59.68 | 59.68 | 3.42 |
| Fungus | Compound | ||
|---|---|---|---|
| TQ | ATQ | Nystatin | |
| MIC (mM) | |||
| C. albicans | 3.81 | 1.75 | 2.70 |
| S. cerevisiae | 1.90 | 0.22 | 1.35 |
| A. braziliensis | 3.81 | 0.88 | 1.35 |
| Compound | Cell Line | ||||
|---|---|---|---|---|---|
| SW620 | CFPAC | HepG2 | HeLa | WI38 | |
| IC50 (µM) | |||||
| TQ | 39.95 | 48.26 | 23.97 | 28.85 | 19.83 |
| ATQ | 25.82 | 30.87 | 8.26 | 15.74 | 9.41 |
| 1a | >100 | 90.22 | 39.34 | 32.22 | 19.89 |
| 1b | 43.68 | 31.02 | 32.27 | 25.00 | 29.81 |
| 1c | >100 | >100 | >100 | >100 | 84.48 |
| 1d | 51.53 | 34.51 | 30.46 | 30.10 | 57.61 |
| 1e | 79.88 | 49.47 | 39.48 | 42.67 | >100 |
| 1f | 5.82 | 33.81 | 9.36 | 4.13 | 5.54 |
| 1g | 54.93 | >100 | >100 | >100 | 6.21 |
| 1h | >100 | 97.00 | >100 | >100 | 75.89 |
| 1i | 55.30 | 40.58 | 4.58 | 6.38 | 9.32 |
| 1j | 31.55 | 27.01 | 7.28 | 10.27 | 9.97 |
| Compound | PTEN | Topoisomerase II | NFκB | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) | Ki * Value (mM) | No. of H bonds | Binding Energy (kcal/mol) | Ki * Value (mM) | No. of H bonds | Binding Energy (kcal/mol) | Ki * Value (mM) | No. of H bonds | |
| TQ | −4.02 | 1.12 | 1 | −3.02 | 6.1 | 0 | −4.75 | 0.328 | 1 |
| 1e | −3.57 | 2.41 | 0 | −4.32 | 0.677 | 0 | −6.02 | 0.039 | 2 |
| 1f | −3.88 | 1.43 | 0 | −3.80 | 1.65 | 0 | −5.58 | 0.082 | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glamočlija, U.; Padhye, S.; Špirtović-Halilović, S.; Osmanović, A.; Veljović, E.; Roca, S.; Novaković, I.; Mandić, B.; Turel, I.; Kljun, J.; et al. Synthesis, Biological Evaluation and Docking Studies of Benzoxazoles Derived from Thymoquinone. Molecules 2018, 23, 3297. https://doi.org/10.3390/molecules23123297
Glamočlija U, Padhye S, Špirtović-Halilović S, Osmanović A, Veljović E, Roca S, Novaković I, Mandić B, Turel I, Kljun J, et al. Synthesis, Biological Evaluation and Docking Studies of Benzoxazoles Derived from Thymoquinone. Molecules. 2018; 23(12):3297. https://doi.org/10.3390/molecules23123297
Chicago/Turabian StyleGlamočlija, Una, Subhash Padhye, Selma Špirtović-Halilović, Amar Osmanović, Elma Veljović, Sunčica Roca, Irena Novaković, Boris Mandić, Iztok Turel, Jakob Kljun, and et al. 2018. "Synthesis, Biological Evaluation and Docking Studies of Benzoxazoles Derived from Thymoquinone" Molecules 23, no. 12: 3297. https://doi.org/10.3390/molecules23123297
APA StyleGlamočlija, U., Padhye, S., Špirtović-Halilović, S., Osmanović, A., Veljović, E., Roca, S., Novaković, I., Mandić, B., Turel, I., Kljun, J., Trifunović, S., Kahrović, E., Kraljević Pavelić, S., Harej, A., Klobučar, M., & Završnik, D. (2018). Synthesis, Biological Evaluation and Docking Studies of Benzoxazoles Derived from Thymoquinone. Molecules, 23(12), 3297. https://doi.org/10.3390/molecules23123297








