Expression Analysis of Oxalate Metabolic Pathway Genes Reveals Oxalate Regulation Patterns in Spinach
Abstract
:1. Introduction
2. Results
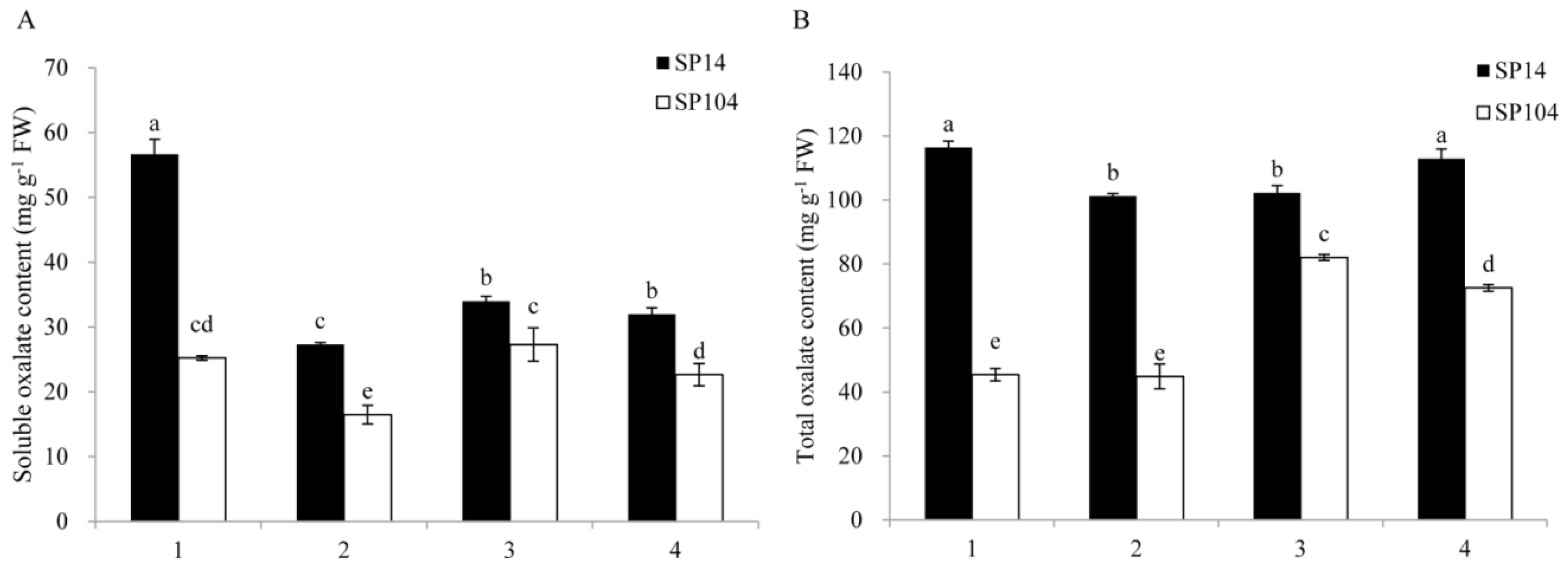
2.1. Oxalate Accumulation under Normal Growth Conditions in Spinach
2.2. Oxalate Accumulation under NO3− and NH4+ Growth Conditions in Spinach
2.3. Identification of Putative Oxalate Biosynthesis and Metabolism Genes
2.4. Expression of Putative Genes Involved in Oxalate Biosynthesis in Spinach
2.5. Expression of Putative Oxalate Degradation Genes in Spinach
2.6. Expression of Putative Oxalate Related Genes under NO3− and NH4+ Treatments
3. Discussion
4. Materials and Methods
4.1. Materials and Treatments
4.2. Determination of Oxalate Levels
4.3. Identification of Putative Oxalate Biosynthesis and Metabolism Genes
4.4. RNA Extraction and Real-Time qRT-PCR Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van Treuren, R.; Coquin, P.; Lohwasser, U. Genetic resources collections of leafy vegetables (lettuce, spinach, chicory, artichoke, asparagus, lamb’s lettuce, rhubarb and rocket salad): Composition and gaps. Genet. Resour. Crop Evol. 2011, 59, 981–997. [Google Scholar] [CrossRef]
- Correll, J.C.; Bluhm, B.H.; Feng, C.; Lamour, K.; du Toit, L.J.; Koike, S.T. Spinach: Better management of downy mildew and white rust through genomics. Eur. J. Plant Pathol. 2011, 129, 193–205. [Google Scholar] [CrossRef]
- Lester, G.E.; Makus, D.J.; Hodges, D.M.; Jifon, J.L. Summer (Subarctic) versus winter (Subtropic) production affects spinach (Spinacia oleracea L.) leaf bionutrients: Vitamins (C, E, folate, K1, provitamin A), lutein, phenolics, and antioxidants. J. Agric. Food Chem. 2013, 61, 7019–7027. [Google Scholar] [CrossRef] [PubMed]
- Mou, B.Q. Evaluation of oxalate concentration in the U.S. spinach germplasm collection. Hortscience 2008, 43, 1690–1693. [Google Scholar]
- Kaminishi, A.; Kita, N. Seasonal change of nitrate and oxalate concentration in relation to the growth rate of spinach cultivars. Hortscience 2006, 41, 1589–1595. [Google Scholar]
- Jaworska, G. Content of nitrates, nitrites, and oxalates in New Zealand spinach. Food Chem. 2005, 89, 235–242. [Google Scholar] [CrossRef]
- Kawazu, Y.; Okimura, M.; Ishii, T.; Yui, S. Varietal and seasonal differences in oxalate content of spinach. Sci. Hortic. (Amsterdam) 2003, 97, 203–210. [Google Scholar] [CrossRef]
- Shi, A.N.; Mou, B.Q.; Correll, J.C. Association analysis for oxalate concentration in spinach. Euphytica 2016, 212, 17–28. [Google Scholar] [CrossRef]
- Faheed, F.; Mazen, A.; Abd Elmohsen, S. Physiological and ultrastructural studies on calcium oxalate crystal formation in some plants. Turk. J. Bot. 2013, 37, 139–152. [Google Scholar]
- Korth, K.L.; Doege, S.J.; Park, S.H.; Goggin, F.L.; Wang, Q.; Gomez, S.K.; Liu, G.; Jia, L.; Nakata, P.A. Medicago truncatula mutants demonstrate the role of plant calcium oxalate crystals as an effective defense against chewing insects. Plant Physiol. 2006, 141, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Cunha, W.G.; Tinoco, M.L.P.; Pancoti, H.L.; Ribeiro, R.E.; Aragão, F.J.L. High resistance to Sclerotinia sclerotiorum in transgenic soybean plants transformed to express an oxalate decarboxylase gene. Plant Pathol. 2010, 59, 654–660. [Google Scholar] [CrossRef]
- Franceschi, V.R.; Nakata, P.A. Calcium oxalate in plants: Formation and function. Annu. Rev. Plant Biol. 2005, 56, 41–71. [Google Scholar] [CrossRef] [PubMed]
- Nakata, P.A. Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Sci. 2003, 164, 901–909. [Google Scholar] [CrossRef]
- Zhang, B.; Oakes, A.D.; Newhouse, A.E.; Baier, K.M.; Maynard, C.A.; Powell, W.A. A threshold level of oxalate oxidase transgene expression reduces Cryphonectria parasitica-induced necrosis in a transgenic American chestnut (Castanea dentata) leaf bioassay. Transgenic Res. 2013, 22, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Nakata, P.A. Engineering calcium oxalate crystal formation in Arabidopsis. Plant Cell Physiol. 2012, 53, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Kuo-Huang, L.L.; Ku, M.S.; Franceschi, V.R. Correlations between calcium oxalate crystals and photosynthetic activities in palisade cells of shadeadapted Peperomia glabella. Bot. Stud. 2007, 48, 155–164. [Google Scholar]
- Tooulakou, G.; Giannopoulos, A.; Nikolopoulos, D.; Bresta, P.; Dotsika, E.; Orkoula, M.G.; Kontoyannis, C.G.; Fasseas, C.; Liakopoulos, G.; Klapa, M.I.; et al. Alarm photosynthesis: Calcium oxalate crystals as an internal CO2 source in plants. Plant Physiol. 2016, 171, 2577–2585. [Google Scholar] [CrossRef] [PubMed]
- Klug, B.; Horst, W.J. Oxalate exudation into the root-tip water free space confers protection from aluminum toxicity and allows aluminum accumulation in the symplast in buckwheat (Fagopyrum esculentum). New Phytol. 2010, 187, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Noonan, S.C.; Savage, G.P. Oxalate content of foods and its effect on humans. Asia Pac. J. Clin. Nutr. 1999, 8, 64–74. [Google Scholar] [PubMed]
- Holmes, R.P.; Goodman, H.O.; Assimos, D.G. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int. 2001, 59, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.P.; Kennedy, M. Estimation of the oxalate content of foods and daily oxalate intake. Kidney Int. 2000, 57, 1662–1667. [Google Scholar] [CrossRef] [PubMed]
- Bong, W.C.; Vanhanen, L.P.; Savage, G.P. Addition of calcium compounds to reduce soluble oxalate in a high oxalate food system. Food Chem. 2017, 221, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, X.; Zhang, Y.; Zheng, S.J.; Du, S. Effects of nitrogen levels and nitrate/ammonium ratios on oxalate concentrations of different forms in edible parts of spinach. J. Plant Nutr. 2005, 28, 2011–2025. [Google Scholar] [CrossRef]
- Liu, X.X.; Lu, L.L.; Chen, Q.H.; Ding, W.Y.; Dai, P.B.; Hu, Y.; Yu, Y.; Jin, C.W.; Lin, X.Y. Ammonium reduces oxalate accumulation in different spinach (Spinacia oleracea L.) genotypes by inhibiting root uptake of nitrate. Food Chem. 2015, 186, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Moscatello, S.; Leccese, A.; Colla, G.; Battistelli, A. The effect of growing spinach (Spinacia oleracea L.) at two light intensities on the amounts of oxalate, ascorbate and nitrate in their leaves. J. Hortic. Sci. Biotechnol. 2004, 79, 606–609. [Google Scholar] [CrossRef]
- Lin, X.Y.; Liu, X.X.; Zhang, Y.P.; Zhou, Y.Q.; Hu, Y.; Chen, Q.H.; Zhang, Y.S.; Jin, C.W. Short-term alteration of nitrogen supply prior to harvest affects quality in hydroponic-cultivated spinach (Spinacia oleracea). J. Sci. Food Agric. 2014, 94, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Al Daini, H.; Norman, H.C.; Young, P.; Barrett-Lennard, E.G. The source of nitrogen (NH4+ or NO3−) affects the concentration of oxalate in the shoots and the growth of Atriplex nummularia (oldman saltbush). Funct. Plant Biol. 2013, 40, 1057–1064. [Google Scholar] [CrossRef]
- Libert, B.; Franceschi, V.R. Oxalate in crop plants. J. Agric. Food Chem. 1987, 35, 926–938. [Google Scholar] [CrossRef]
- Palaniswamy, U.R.; Bible, B.B.; McAvoy, R.J. Effect of nitrate: Ammonium nitrogen ratio on oxalate levels of purslane. Trends New Crops New Uses 2002, 11, 453–455. [Google Scholar]
- Cai, X.F.; Xu, C.X.; Wang, X.L.; Ge, C.H.; Wang, Q.H. The oxalic acid in plants: Biosynthesis, degradation and its accumulation regulation (Review in Chinese). Plant Physiol. J. 2015, 51, 267–272. [Google Scholar]
- Richardson, K.E.; Tolbert, N.E. Oxidation of glyoxylic acid to oxalic acid by glycolic acid oxidase. J. Biol. Chem. 1961, 236, 1280–1284. [Google Scholar] [PubMed]
- Ishikawa, T.; Dowdle, J.; Smirnoff, N. Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiol. Plant 2006, 126, 343–355. [Google Scholar] [CrossRef]
- Fujii, N.; Watanabe, M.; Watanabe, Y.; Shimada, N. Kate of oxalate biosynthesis from glycolate and ascorbic acid in spinach leaves. Soil Sci. Plant Nutr. 1993, 39, 627–634. [Google Scholar] [CrossRef]
- Yu, L.; Jiang, J.Z.; Zhang, C.; Jiang, L.R.; Ye, N.H.; Lu, Y.S.; Yang, G.Z.; Liu, E.; Peng, C.L.; He, Z.H.; et al. Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 2010, 61, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Beevers, H. Biogenesis of oxalate in plant tissues. Plant Physiol. 1968, 43, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Kostman, T.A.; Tarlyn, N.M.; Loewus, F.A.; Franceschi, V.R. Biosynthesis of L-ascorbic acid and conversion of carbons 1 and 2 of l-ascorbic acid to oxalic acid occurs within individual calcium oxalate crystal idioblasts. Plant Physiol. 2001, 125, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Loewus, F.A. Metabolic conversion of L-ascorbic acid to oxalic acid in oxalate-accumulating plants. Plant Physiol. 1975, 56, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Keates, S.E.; Tarlyn, N.M.; Loewus, F.A.; Franceschi, V.R. L-ascorbic acid and l-galactose are sources for oxalic acid and calcium oxalate in Pistia stratiotes. Phytochemistry 2000, 53, 433–440. [Google Scholar] [CrossRef]
- Li, W.; Li, W.X.; Jia, L.; Gang, Y.Z.; Min, D.Z. Research on oxalate oxidase and its genes in plants. Agric. Sci. Technol. 2011, 12, 11–13, 19. [Google Scholar]
- Svedruzic, D.; Jonsson, S.; Toyota, C.G.; Reinhardt, L.A.; Ricagno, S.; Lindqvist, Y.; Richards, N.G. The enzymes of oxalate metabolism: Unexpected structures and mechanisms. Arch. Biochem. Biophys. 2005, 433, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.; Kim, H.U.; Nakata, P.A.; Browse, J. A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell. 2012, 24, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.X.; Jiao, C.; Zheng, Y.; Sun, H.H.; Liu, W.L.; Cai, X.F.; Wang, X.L.; Liu, S.; Xu, Y.M.; Mou, B.Q.; et al. De novo and comparative transcriptome analysis of cultivated and wild spinach. Sci. Rep. (UK) 2015, 5, 17706. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.X.; Jiao, C.; Sun, H.H.; Cai, X.F.; Wang, X.L.; Ge, C.H.; Zheng, Y.; Liu, W.L.; Sun, X.P.; Xu, Y.M.; et al. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 2017, 8, 15275. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Joosten, H.J.; Niu, W.; Zhao, Z.; Mariano, P.S.; McCalman, M.; van Kan, J.; Schaap, P.J.; Dunaway-Mariano, D. Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi. J. Biol. Chem. 2007, 282, 9581–9590. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Min, J.Y.; Dickman, M.B. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. MPMI 2008, 21, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Errakhi, R.; Meimoun, P.; Lehner, A.; Vidal, G.; Briand, J.; Corbineau, F.; Rona, J.P.; Bouteau, F. Anion channel activity is necessary to induce ethylene synthesis and programmed cell death in response to oxalic acid. J. Exp. Bot. 2008, 59, 3121–3129. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, N.; Ghosh, R.; Ghosh, S.; Narula, K.; Tayal, R.; Datta, A.; Chakraborty, S. Reduction of oxalate levels in tomato fruit and consequent metabolic remodeling following overexpression of a fungal oxalate decarboxylase. Plant Physiol. 2013, 162, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Walz, A.; Zingen-Sell, I.; Loeffler, M.; Sauer, M. Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathol. 2008, 57, 453–458. [Google Scholar] [CrossRef]
- Ji, X.M.; Peng, X.X. Oxalate accumulation as regulated by nitrogen forms and its relationship to photosynthesis in rice (Oryza sativa L.). J. Integr. Plant Biol. 2005, 47, 831–838. [Google Scholar] [CrossRef]
- Cai, X.F.; Zhang, Y.Y.; Zhang, C.J.; Zhang, T.Y.; Hu, T.X.; Ye, J.; Zhang, J.H.; Wang, T.T.; Li, H.X.; Ye, Z.B. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |






| Num | Gene Name | Unigene Number | Chomosome/Scaffold and Location | Strand | CDS (bp) | Size (aa) | Introns | ||
|---|---|---|---|---|---|---|---|---|---|
| 1 | SoGLO1 | Spo19861 | chr4 | 118492560 | 118496915 | Rev | 1053 | 350 | 10 |
| 2 | SoGLO2 | Spo21903 | chr5 | 35418377 | 35422331 | For | 1107 | 368 | 10 |
| 3 | SoGLO3 | Spo10076 | chr1 | 45522012 | 45524813 | Rev | 1095 | 364 | 9 |
| 4 | SoGLO4 | Spo21282 | SpoScf_01013 | 111890 | 113703 | For | 756 | 251 | 7 |
| 5 | SoGLO5 | Spo20781 | chr4 | 118492935 | 118496947 | Rev | 720 | 239 | 7 |
| 6 | SoOXAC1 | Spo00571 | SpoScf_01500 | 31009 | 43616 | For | 1875 | 624 | 9 |
| 7 | SoOXAC2 | Spo21624 | chr2 | 55761708 | 55765402 | Rev | 909 | 302 | 4 |
| 8 | SoOXAC3 | Spo21589 | chr2 | 55798280 | 55800798 | For | 915 | 304 | 4 |
| 9 | SoMLS | Spo16696 | chr5 | 17999221 | 18002655 | For | 1713 | 570 | 3 |
| 10 | SoMDH1 | Spo21995 | SpoScf_02896 | 22145 | 23389 | For | 1245 | 414 | 0 |
| 11 | SoMDH2 | Spo08175 | chr5 | 10849769 | 10851010 | Rev | 1242 | 413 | 0 |
| 12 | SoMDH3 | Spo10516 | chr4 | 62141844 | 62145553 | For | 1032 | 343 | 6 |
| 13 | SoMDH4 | Spo22090 | SpoScf_03526 | 16484 | 21730 | Rev | 1077 | 358 | 7 |
| 14 | SoCTS1 | Spo11084 | chr4 | 101756403 | 101770489 | Rev | 1800 | 599 | 19 |
| 15 | SoCTS2 | Spo11913 | chr4 | 7539409 | 7552423 | For | 1608 | 535 | 20 |
| 16 | SoACO | Spo13736 | SpoScf_03007 | 28088 | 36725 | Rev | 2967 | 988 | 18 |
| 17 | SoICL | Spo13898 | SpoScf_00215 | 416756 | 427351 | For | 1989 | 662 | 6 |
| 18 | SoAAE3 | Spo04424 | chr3 | 3466032 | 3471417 | For | 1575 | 524 | 3 |
| 19 | SoOXO1 | Spo14475 | chr2 | 40519401 | 40522642 | Rev | 666 | 221 | 1 |
| 20 | SoOXO2 | Spo04401 | chr3 | 2862241 | 2863645 | For | 654 | 217 | 1 |
| 21 | SoOXDE | Spo00223 | chr1 | 31229950 | 31233690 | Rev | 1722 | 573 | 1 |
| 22 | SoFXDE | Spo19843 | chr4 | 119024460 | 119033349 | Rev | 1911 | 636 | 8 |
| 23 | SoOXDC1 | Spo06441 | chr4 | 49505283 | 49512025 | For | 1512 | 503 | 4 |
| 24 | SoOXDC2 | Spo19759 | chr5 | 17648569 | 17649072 | For | 504 | 167 | 0 |
| 25 | SoOXDC3 | Spo25084 | chr4 | 4971997 | 4972967 | Rev | 441 | 146 | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, X.; Ge, C.; Xu, C.; Wang, X.; Wang, S.; Wang, Q. Expression Analysis of Oxalate Metabolic Pathway Genes Reveals Oxalate Regulation Patterns in Spinach. Molecules 2018, 23, 1286. https://doi.org/10.3390/molecules23061286
Cai X, Ge C, Xu C, Wang X, Wang S, Wang Q. Expression Analysis of Oxalate Metabolic Pathway Genes Reveals Oxalate Regulation Patterns in Spinach. Molecules. 2018; 23(6):1286. https://doi.org/10.3390/molecules23061286
Chicago/Turabian StyleCai, Xiaofeng, Chenhui Ge, Chenxi Xu, Xiaoli Wang, Shui Wang, and Quanhua Wang. 2018. "Expression Analysis of Oxalate Metabolic Pathway Genes Reveals Oxalate Regulation Patterns in Spinach" Molecules 23, no. 6: 1286. https://doi.org/10.3390/molecules23061286
APA StyleCai, X., Ge, C., Xu, C., Wang, X., Wang, S., & Wang, Q. (2018). Expression Analysis of Oxalate Metabolic Pathway Genes Reveals Oxalate Regulation Patterns in Spinach. Molecules, 23(6), 1286. https://doi.org/10.3390/molecules23061286





