QSAR Study on Antioxidant Tripeptides and the Antioxidant Activity of the Designed Tripeptides in Free Radical Systems
Abstract
:1. Introduction
2. Results and Discussions
2.1. QSAR Modeling and Predominant Residue Analysis
2.2. Determination of Relative Antioxidant Activity
2.2.1. DPPH Radical Scavenging Capacity
2.2.2. Trolox Equivalent Antioxidant Capacity (TEAC) Assay
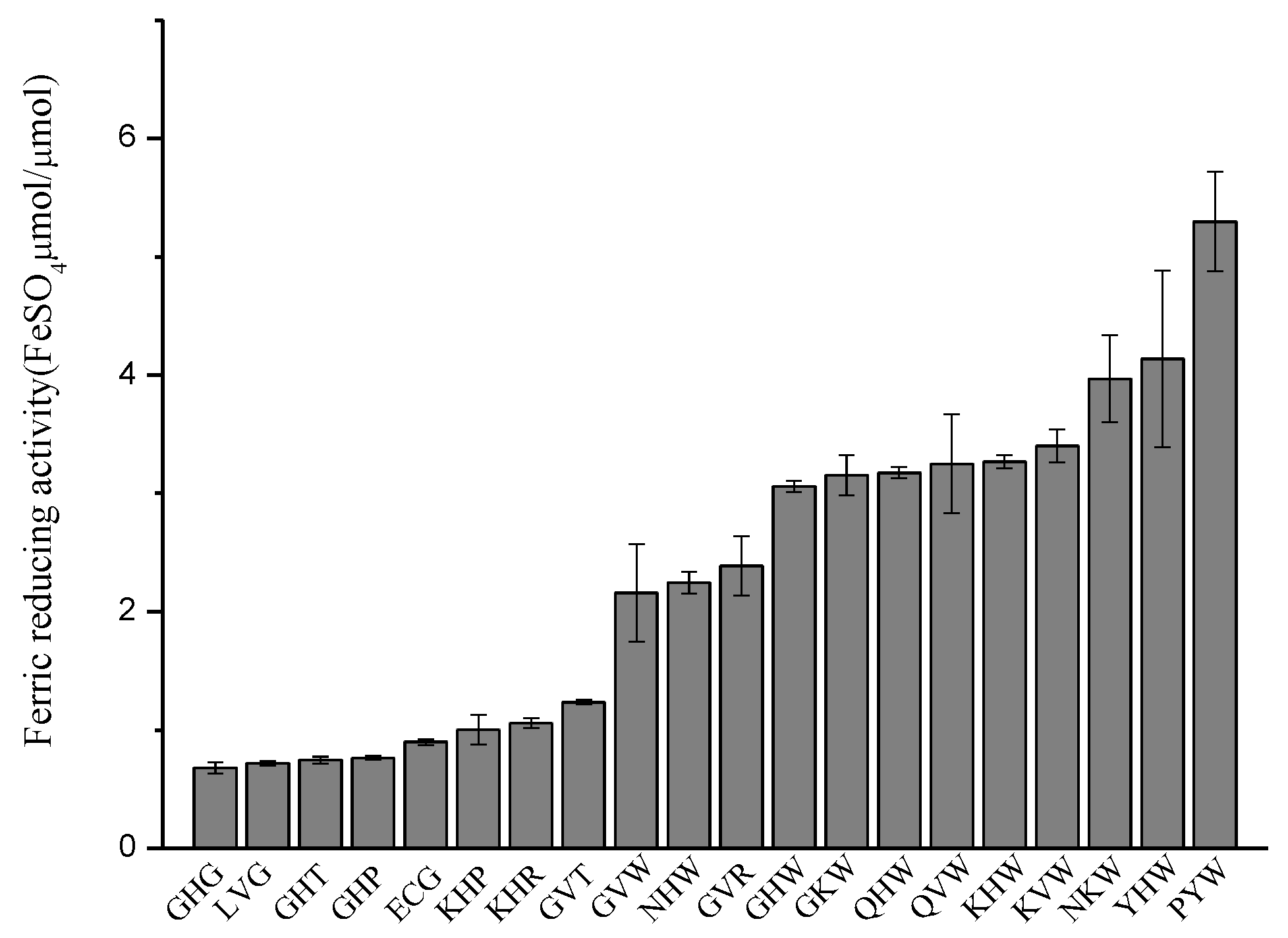
2.2.3. Ferric Reducing Antioxidant Power (FRAP) Assay
2.3. Validation of QSAR Model
3. Materials and Methods
3.1. Data Source
3.2. Structural Characterization of Tripeptides
3.3. Quantitative Structure-Activity Relationship Modeling
3.4. Rational Design of Antioxidant Peptides
3.5. Antioxidant Activity Assay
3.5.1. DPPH Radical Scavenging Activities
3.5.2. Trolox Equivalent Antioxidant Capacity (TEAC) Assay
3.5.3. Ferric Reducing Antioxidant Power (FRAP) Assay
3.6. Statistical Analysis
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simões, J.; Moreira, A.S.P.; da Costa, E.; Evtyugin, D.; Domingues, P.; Nunes, F.M.; Coimbra, M.A.; Domingues, M.R.M. Oxidation of amylose and amylopectin by hydroxyl radicals assessed by electrospray ionisation mass spectrometry. Carbohydr. Polym. 2016, 148, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Van den Ende, W.; Peshev, D.; De Gara, L. Disease prevention by natural antioxidants and prebiotics acting as ROS scavengers in the gastrointestinal tract. Trends Food Sci. Technol. 2011, 22, 689–697. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Castegna, A.; Pocernich, C.B.; Drake, J.; Scapagnini, G.; Calabrese, V. Nutritional approaches to combat oxidative stress in Alzheimer’s disease. J. Nutr. Biochem. 2002, 13, 444–461. [Google Scholar] [CrossRef] [Green Version]
- Kohen, R.; Yamamoto, Y.; Cundy, K.C.; Ames, B.N. Antioxidant activity of carnosine, homocarnosine, and anserine present in muscle and brain. Proc. Natil. Acad. Sci. USA 1988, 85, 3175–3179. [Google Scholar] [CrossRef]
- Gu, L.; Zhao, M.; Li, W.; You, L.; Wang, J.; Wang, H.; Ren, J. Chemical and cellular antioxidant activity of two novel peptides designed based on glutathione structure. Food Chem. Toxicol. 2012, 50, 4085–4091. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ding, L.; Yu, Z.; Zhang, T.; Ma, S.; Liu, J. Intracellular ROS scavenging and antioxidant enzyme regulating capacities of corn gluten meal-derived antioxidant peptides in HepG2 cells. Food Res. Int. 2016, 90, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.-J.; Jung, W.-K.; Kim, S.-K. Free radical scavenging activity of a novel antioxidative peptide purified from hydrolysate of bullfrog skin, Rana catesbeiana Shaw. Bioresour. Technol. 2008, 99, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Jacinto-Hernández, C.; Alaiz, M.; Girón-Calle, J.; Vioque, J.; Dávila-Ortiz, G. Antioxidant and metal chelating activities of peptide fractions from phaseolin and bean protein hydrolysates. Food Chem. 2012, 135, 1789–1795. [Google Scholar] [CrossRef] [PubMed]
- Megías, C.; Pedroche, J.; Yust, M.M.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Production of copper-chelating peptides after hydrolysis of sunflower proteins with pepsin and pancreatin. LWT-Food Sci. Technol. 2008, 41, 1973–1977. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, T.; Jiang, B.; Miao, M.; Mu, W. The effects of an antioxidative pentapeptide derived from chickpea protein hydrolysates on oxidative stress in Caco-2 and HT-29 cell lines. J. Funct. Foods 2014, 7, 719–726. [Google Scholar] [CrossRef]
- Shi, Y.; Kovacs-Nolan, J.; Jiang, B.; Tsao, R.; Mine, Y. Peptides derived from eggshell membrane improve antioxidant enzyme activity and glutathione synthesis against oxidative damage in Caco-2 cells. J. Funct. Foods 2014, 11, 571–580. [Google Scholar] [CrossRef]
- Kerasioti, E.; Stagos, D.; Tzimi, A.; Kouretas, D. Increase in antioxidant activity by sheep/goat whey protein through nuclear factor-like 2 (Nrf2) is cell type dependent. Food Chem. Toxicol. 2016, 97, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Nikoo, M.; Benjakul, S. Potential application of seafood-derived peptides as bifunctional ingredients, antioxidant–cryoprotectant: A review. J. Funct. Foods 2015, 19, 753–764. [Google Scholar] [CrossRef]
- Zou, T.-B.; He, T.-P.; Li, H.-B.; Tang, H.-W.; Xia, E.-Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhao, Y.; Dong, H.; Su, G.; Zhao, M. Structure–activity relationship of antioxidant dipeptides: Dominant role of Tyr, Trp, Cys and Met residues. J. Funct. Foods 2016, 21, 485–496. [Google Scholar] [CrossRef]
- Chen, H.-M.; Muramoto, K.; Yamauchi, F.; Nokihara, K. Antioxidant Activity of Designed Peptides Based on the Antioxidative Peptide Isolated from Digests of a Soybean Protein. J. Agric. Food Chem. 1996, 44, 2619–2623. [Google Scholar] [CrossRef]
- Saito, K.; Jin, D.; Ogawa, T.; Muramoto, K.; Hatakeyama, E.; Yasuhara, T.; Nokihara, K. Antioxidative properties of tripeptide libraries perpared by the combinatorial chemistry. J. Agric. Food Chem. 2003, 51, 3668–3674. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-W.; Li, B.; He, J.; Qian, P. Quantitative structure–activity relationship study of antioxidative peptide by using different sets of amino acids descriptors. J. Mol. Struct. 2011, 998, 53–61. [Google Scholar] [CrossRef]
- Li, Y.-W.; Li, B. Characterization of structure–antioxidant activity relationship of peptides in free radical systems using QSAR models: Key sequence positions and their amino acid properties. J. Theor. Biol. 2013, 318, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Protasiewicz, M.; Mogut, D. Chemometrics and cheminformatics in the analysis of biologically active peptides from food sources. J. Funct. Foods 2015, 16, 334–351. [Google Scholar] [CrossRef]
- Meng, D.; Zhang, P.; Zhang, L.; Wang, H.; Ho, C.-T.; Li, S.; Shahidi, F.; Zhao, H. Detection of cellular redox reactions and antioxidant activity assays. J. Funct. Foods 2017, 37, 467–479. [Google Scholar] [CrossRef]
- Meng, D.; Zhang, P.; Li, S.; Ho, C.-T.; Zhao, H. Antioxidant activity evaluation of dietary phytochemicals using Saccharomyces cerevisiae as a model. J. Funct. Foods 2017, 38, 36–44. [Google Scholar] [CrossRef]
- Levitt, M. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976, 104, 59–107. [Google Scholar] [CrossRef]
- Cohn, E.J.; Edsall, J.T. Proteins, amino acids and peptides as ions and dipolar ions; Reinhold Publishing Corporation: New York, NY, USA, 1943. [Google Scholar]
- Charton, M.; Charton, B.I. The structural dependence of amino acid hydrophobicity parameters. J. Theor. Biol. 1982, 99, 629–644. [Google Scholar] [CrossRef]
- FAUCHÈRE, J.L.; Charton, M.; Kier, L.B.; Verloop, A.; Pliska, V. Amino acid side chain parameters for correlation studies in biology and pharmacology. Int. J. Pept. Protein Res. 1988, 32, 269–278. [Google Scholar]
- Qian, N.; Sejnowski, T.J. Predicting the secondary structure of globular proteins using neural network models. J. Mol. Biol. 1988, 202, 865–2884. [Google Scholar] [CrossRef]
- Yutani, K.; Ogasahara, K.; Tsujita, T.; Sugino, Y. Dependence of conformational stability on hydrophobicity of the amino acid residue in a series of variant proteins substituted at a unique position of tryptophan synthase alpha subunit. Proc. Nat. Acad. Sci. USA 1987, 84, 4441–4444. [Google Scholar] [CrossRef] [PubMed]
- Oobatake, M.; Kubota, Y.; Ooi, T. Optimization of Amino Acid Parameters for Correspondence of Sequence to Tertiary Structures of Proteins. Bull. Inst. Chem. Res. 1985, 63, 84–92. [Google Scholar]
- Agrawal, H.; Joshi, R.; Gupta, M. Isolation, purification and characterization of antioxidative peptide of pearl millet (Pennisetum glaucum) protein hydrolysate. Food Chem. 2016, 204, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, X.; Lin, S.; Zhang, Z.; Chen, F. Identification of novel peptides from 3 to 10kDa pine nut (Pinus koraiensis) meal protein, with an exploration of the relationship between their antioxidant activities and secondary structure. Food Chem. 2017, 219, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Lin, L.; Su, G.; Zhao, Q.; Zhao, M. Pitfalls of using 1,1-diphenyl-2-picrylhydrazyl (DPPH) assay to assess the radical scavenging activity of peptides: Its susceptibility to interference and low reactivity towards peptides. Food Res. Int. 2015, 76, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Matsui, R.; Honda, R.; Kanome, M.; Hagiwara, A.; Matsuda, Y.; Togitani, T.; Ikemoto, N.; Terashima, M. Designing antioxidant peptides based on the antioxidant properties of the amino acid side-chains. Food Chem. 2018, 245, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Kouzuma, Y.; Yonekura, M. Structures and properties of antioxidative peptides derived from royal jelly protein. Food Chem. 2009, 113, 238–245. [Google Scholar] [CrossRef]
- Luo, J.; Li, L.; Kong, L. Preparative separation of phenylpropenoid glycerides from the bulbs of Lilium lancifolium by high-speed counter-current chromatography and evaluation of their antioxidant activities. Food Chem. 2012, 131, 1056–1062. [Google Scholar] [CrossRef]
- Kawashima, S.; Pokarowski, P.; Pokarowska, M.; Kolinski, A.; Katayama, T.; Kanehisa, M. AAindex: Amino acid index database, progress report 2008. Nucleic Acids Res. 2007, 36, D202–D205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, Y.; Wen, H.; Lin, Y.; Hu, Y.; Zhang, Y.; Xia, Q.; Lin, Z. QSAR modeling and design of cationic antimicrobial peptides based on structural properties of amino acids. Comb. Chem. High Throughput Screen. 2012, 15, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Lin, Y.; Wen, H.; Wang, L.; Xia, Q.; Lin, Z. TAP-Binding Peptides Prediction by QSAR Modeling Based on Amino Acid Structural Information. Curr. Comput.-Aided Drug Des. 2012, 8, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, Y.; Shu, M.; Wang, R.; Hu, Y.; Lin, Z. Proteasomal cleavage site prediction of protein antigen using BP neural network based on a new set of amino acid descriptor. J. Mol. Model. 2013, 19, 3045–3052. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, P.; Lin, Y.; Shu, M.; Hu, Y.; Xia, Q.; Lin, Z. Quantitative prediction of class I MHC/epitope binding affinity using QSAR modeling derived from amino acid structural information. Comb. Chem. High Throughput Screen. 2015, 18, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Garg, P.; Roy, N. Identification of Novel HIV-1 Integrase Inhibitors Using Shape-Based Screening, QSAR, and Docking Approach. Chem. Boil. Drug Des. 2012, 79, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Deswal, S.; Roy, N. Quantitative structure activity relationship studies of aryl heterocycle-based thrombin inhibitors. Eur. J. Med. Chem. 2006, 41, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Musa, K.H.; Abdullah, A.; Al-Haiqi, A. Determination of DPPH free radical scavenging activity: Application of artificial neural networks. Food Chem. 2016, 194, 705–711. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the 19 tripeptides are available from the authors. |





| Name | Description |
|---|---|
| LEVM760107 | van der Waals parameter [24] |
| COHE430101 | Partial specific volume [25] |
| CHAM820102 | Free energy of solution in water, kcal/mole [26] |
| FAUJ880112 | Negative charge [27] |
| QIAN880115 | Weights for beta-sheet at the window position of -5 [28] |
| YUTK870102 | Unfolding Gibbs energy in water, pH9.0 [29] |
| OOBM850102 | Optimized propensity to form reverse turn [30] |
| Methods | Training Dataset | Test Dataset | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | m | R2train | SD | F | Q2LOO | Q24-fold | Q26-fold | Q210-fold | PRESS | n | R2test | ||
| PLS | 60 | 7 | 0.902 | 0.226 | 68.005 | 0.706 | 0.749 | 0.700 | 0.708 | 7.891 | 31 | 0.888 | |
| SWR-SVM | 60 | 7 | 0.875 | 0.254 | 45.820 | 0.764 | 0.764 | 6.327 | 31 | 0.883 | |||
| SWR-RF | 60 | 7 | 0.868 | 0.261 | 48.857 | 0.728 | 0.728 | 7.298 | 31 | 0.914 | |||
| SWR-MLR | 60 | 7 | 0.893 | 0.235 | 61.745 | 0.798 | 0.792 | 0.802 | 0.822 | 5.413 | 31 | 0.897 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, N.; Chen, J.; Yao, B.; Li, Z. QSAR Study on Antioxidant Tripeptides and the Antioxidant Activity of the Designed Tripeptides in Free Radical Systems. Molecules 2018, 23, 1407. https://doi.org/10.3390/molecules23061407
Chen N, Chen J, Yao B, Li Z. QSAR Study on Antioxidant Tripeptides and the Antioxidant Activity of the Designed Tripeptides in Free Radical Systems. Molecules. 2018; 23(6):1407. https://doi.org/10.3390/molecules23061407
Chicago/Turabian StyleChen, Nan, Ji Chen, Bo Yao, and Zhengguo Li. 2018. "QSAR Study on Antioxidant Tripeptides and the Antioxidant Activity of the Designed Tripeptides in Free Radical Systems" Molecules 23, no. 6: 1407. https://doi.org/10.3390/molecules23061407
APA StyleChen, N., Chen, J., Yao, B., & Li, Z. (2018). QSAR Study on Antioxidant Tripeptides and the Antioxidant Activity of the Designed Tripeptides in Free Radical Systems. Molecules, 23(6), 1407. https://doi.org/10.3390/molecules23061407





