Quantitative Determination of Thiabendazole in Soil Extracts by Surface-Enhanced Raman Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
2.1. The TBZ Molecule and Its Assignment of Raman Peaks
- (1)
- C-H and ring vibration: the vibration of C-H groups’ in-plane and out-of-plane bending deformation concentrated on 700–1400 cm−1 [21]. Figure 1b revealed that the spectral peak intensity at 778, 1011, 1255 and 1277 cm−1 were relatively higher than others, resulting of the strong C-H vibration among aliphatic compounds as well as the widest band range of C-H vibration [19]. Additionally, 1199 cm−1 and 1255 cm−1 belonged to the ring vibration, 1277 cm−1 was the synergistic effects of ring vibration and C-H groups surface vibration. Moreover, 778 cm−1 was the C-H outer surface bending vibration, 1011, 1118, 1303 and 1154 cm−1 belonged to the surface bending vibration of the C-H group [20].
- (2)
- C=N vibration: the vibration of the C=N group is generally in the range of 1300–1700 cm−1, among which the spectral peak intensity at 1456, 1577 and 1591 cm−1 were strong. Refer to Sundaraganesan’s study [19], the C=N stretching vibration band of the ammonium imine group ranges from 1610 to 1690 cm−1, and the Raman peak of C=C is normally weaker than that of the C=N group. Thus, it could be confirmed that 1577, 1591 and 1623 cm−1 belong to the C=N stretching vibration, which was also basically consistent with the vibration peaks of C=N in TBZ identified by Cirak [22].
- (3)
- C-C and C=C vibration: the spectral bands of C-C and C=C are usually in the range of 300–1700 cm−1 and these spectral peaks mainly belong to the characteristic peaks of polycyclic aromatic hydrocarbons in TBZ [21]. Among them, 615 and 632 cm−1 were the synergistic effects of C-C-C and S-C-N inner surface bending and deformable vibration, 876 and 897 cm−1 were the synergistic effects C-C-C outer surface bending deformable vibration and the surface bending vibration of C-H group. Furthermore, 1403 cm−1 belonged to the stretching vibration of C=C band and 1493 cm−1 was the synergistic effects of C=C stretching vibration and N-H inner surface bending vibration. N. Sundaraganesan et al. [19] calculated the vibrational peaks of the C=C group using density functional theory, whose calculation results were consistent with this study as well.
- (4)
- N-H vibration: the N-H band generally belongs to the range of 300–1700 cm−1. In this paper, the N-H in-plane deformation vibration signal appears at 1493 cm−1 and this signal is relatively weak. Reference [23] showed that N-H vibration of aminobenzenes occurred near 1510 cm−1 and this peak was also interfaced with the C=C stretching vibration, which was in accordance with our study.
- (5)
- C-S vibration: the C-S band vibration normally ranges from 300 to 1200 cm−1. Compared with the spectral peak range of the sulfur-containing compounds of C-S group and S-C-N group [24], 985 cm−1 belonged to the C-S stretching vibration, which was consistent with the experimental results obtained by Kim et al. [20].
2.2. Comparison of SERS and Raman Spectrum of TBZ
2.3. Gold Nanoparticles and Its Spectral Analysis
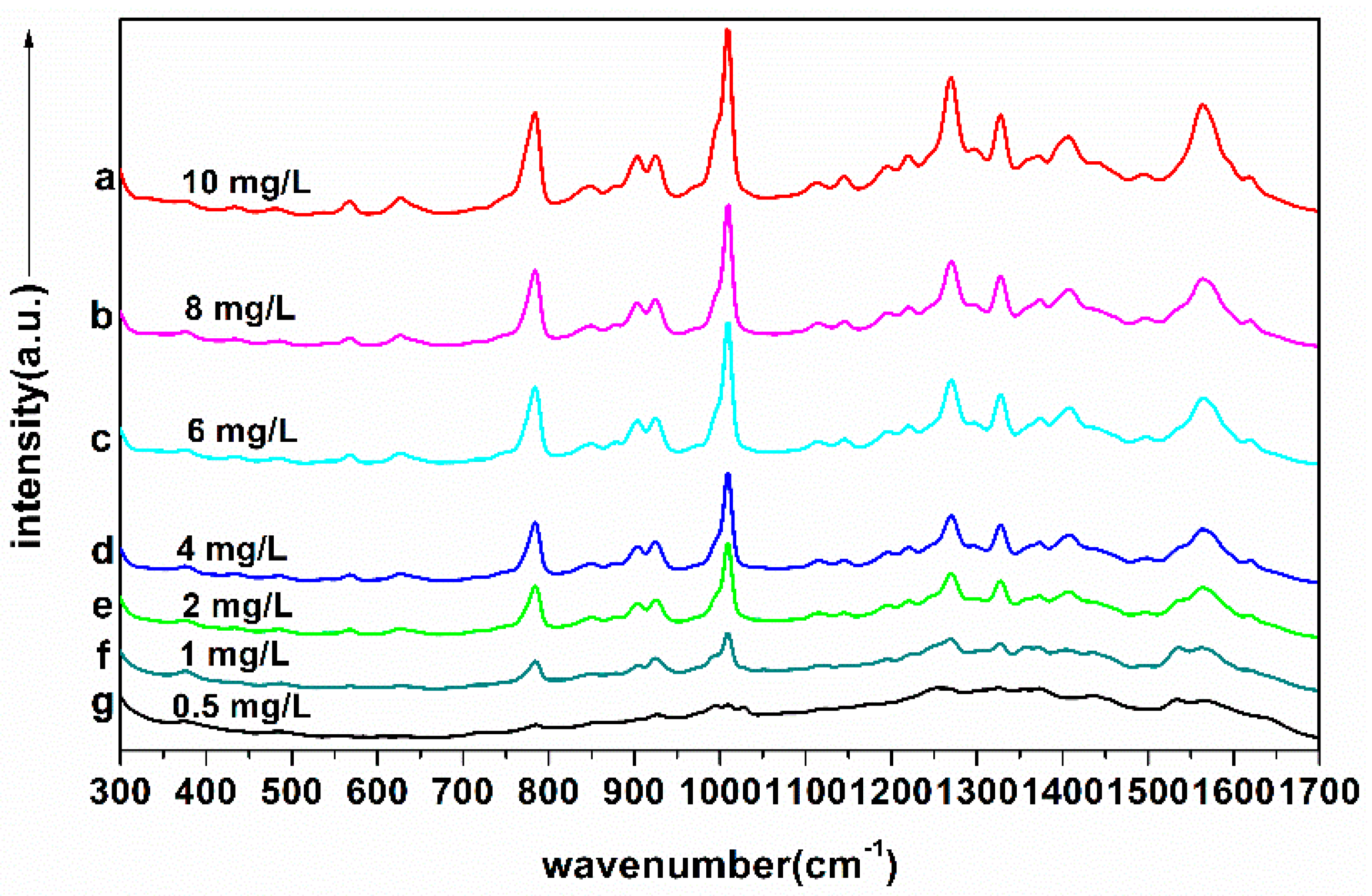
2.4. Determination of Detection Line
2.5. The Linear Regression Equation of TBZ in Red Soil Extracts
2.6. The Analysis of 300–1700 cm−1 SERS of Deltamethrin Pesticides in Soil
2.7. Model Accuracy Verification
3. Materials and Methods
3.1. Experimental Instruments and Reagents
3.2. Experimental Methods
3.3. Gold Nano-Substrate Preparation
3.4. Density Functional Theory (DFT)
3.5. Spectral Preprocessing Methods
3.6. Modeling Method
3.7. Model Evaluation Index
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rodríguez, R.; Picó, Y.; Font, G.; Mañes, J. Analysis of thiabendazole and procymidone in fruits and vegetables by capillary electrophoresis-electrospray mass spectrometry. J. Chromatogr. 2002, 949, 359–366. [Google Scholar] [CrossRef]
- Aharonson, N.; Rubin, B.; Katan, J.; Benjamin, A. Effect of methyl bromide or solar heating treatments on the persistence of pesticides in the soil. In Pesticide Chemistry: Human Welfare and the Environment: Pesticide Residues and Formulation Chemistry; Pergamon Press: Toronto, ON, Canada, 1983; pp. 189–194. [Google Scholar]
- Rong, T.; Yang, R.J.; Christopher, Y.; Zhu, H. Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC). J. Agric. Food Chem. 2003, 51, 6347–6353. [Google Scholar]
- Roessner, U.; Wagner, C.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. Cell Mol. Biol. 2010, 23, 131–142. [Google Scholar] [CrossRef]
- Hugli, T.E.; Moore, S. Determination of the tryptophan content of proteins by ion exchange chromatography of alkaline hydrolysates. J. Biol. Chem. 1972, 247, 2828. [Google Scholar] [PubMed]
- Kasicka, V. Recent developments in capillary electrophoresis and capillary electrochromatography of peptides. Electrophoresis 2006, 27, 142–175. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Aboul-Enein, H.Y. Leaching of triazine pesticides in loamy soil and their determination by reversed phase HPLC. Int. J. Environ. Anal. Chem. 2001, 81, 315–322. [Google Scholar] [CrossRef]
- Li, H.; Lee, H.K. Determination of pesticides in soil by liquid-phase microextraction and gas chromatography–mass spectrometry. J. Chromatogr. A 2004, 1038, 37–42. [Google Scholar]
- Dinelli, G.; Vicari, A.; Brandolini, V. Detection and quantitation of sulfonylurea herbicides in soil at the ppb level by capillary electrophoresis. J. Chromatogr. A 1995, 700, 201–207. [Google Scholar] [CrossRef]
- Önal, A. A review: Current analytical methods for the determination of biogenic amines in foods. Food Chem. 2007, 103, 1475–1486. [Google Scholar] [CrossRef]
- Farquharson, S.; Gift, A.; Shende, C.; Inscore, F.; Ordway, B.; Farquharson, C.; Murren, J. Surface-enhanced Raman spectral measurements of 5-fluorouracil in saliva. Molecules 2008, 13, 2608–2627. [Google Scholar] [CrossRef] [PubMed]
- Doering, W.E.; Nie, S. Single-Molecule and Single-Nanoparticle SERS: Examining the Roles of Surface Active Sites and Chemical Enhancement. J. Phys. Chem. B 2002, 106, 311–317. [Google Scholar] [CrossRef]
- Lin, L.; Wu, R.; Liu, M.; Wang, X.; Yan, L. Surface-enhanced Raman spectroscopy analysis of thiabendazole pesticide. Spectrosc. Spectr. Anal. 2015, 35, 404–408. [Google Scholar]
- Luo, H.; Huang, Y.; Lai, K.; Rasco, B.A.; Fan, Y. Surface-enhanced Raman spectroscopy coupled with gold nanoparticles for rapid detection of phosmet and thiabendazole residues in apples. Food Control 2016, 68, 229–235. [Google Scholar] [CrossRef]
- Feng, J.; Hu, Y.; Grant, E.; Lu, X. Determination of thiabendazole in orange juice using an MISPE-SERS chemosensor. Food Chem. 2018, 239, 816–822. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chen, T.; Labuza, T.P. Recovery and quantitative detection of thiabendazole on apples using a surface swab capture method followed by surface-enhanced Raman spectroscopy. Food Chem. 2014, 148, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Chermette, H. Chemical reactivity indexes in density functional theory. J. Comput. Chem. 2015, 20, 129–154. [Google Scholar] [CrossRef]
- Matysik, J.; Hildebrandt, P.; Smit, K.; Mark, F.; Gärtner, W.; Braslavsky, S.E.; Schaffner, K.; Schrader, B. Raman spectroscopic analysis of isomers of biliverdin dimethyl ester. J. Pharm. Biomed. Anal. 1997, 15, 1319–1324. [Google Scholar] [CrossRef]
- Sundaraganesan, N.; Ilakiamani, S.; Subramani, P.; Joshua, B.D. Comparison of experimental and ab initio HF and DFT vibrational spectra of benzimidazole. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2007, 67, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kim, M.K.; Lee, C.J.; Jung, Y.M.; Lee, M.S. Surface-enhanced Raman spectroscopy of benzimidazolic fungicides: Benzimidazole and thiabendazole. Bull. Korean Chem. Soc. 2009, 30, 2930–2934. [Google Scholar] [CrossRef]
- Zhu, Z.Y.; Gu, R.N.; Lu, T.H. Application of Raman Spectroscopy in Chemistry; Northeastern University Press: Boston, MA, USA, 1998. [Google Scholar]
- Cırak, C.; Koç, N. Molecular structure and effects of intermolecular hydrogen bonding on the vibrational spectrum of trifluorothymine, an antitumor and antiviral agent. J. Mol. Model. 2012, 18, 4453–4464. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.M.; Im, J.W.; Kim, E.R.; Lee, H.; Mu, S.L. Surface-enhanced Raman scattering of ruthenium (ii) complex adsorbed on silver colloid. Bull. Korean Chem. Soc. 2001, 22, 318–320. [Google Scholar]
- Tammer, M.; Sokrates, G. Infrared and Raman characteristic group frequencies: Tables and charts. Colloid Polym. Sci. 2004, 283, 235. [Google Scholar] [CrossRef]
- Dasary, S.S.; Singh, A.K.; Senapati, D.; Yu, H.; Ray, P.C. Gold nanoparticle based label-free SERS probe for ultrasensitive and selective detection of trinitrotoluene. J. Am. Chem. Soc. 2009, 131, 13806–13812. [Google Scholar] [CrossRef] [PubMed]
- Perna, G.; Lasalvia, M.; D’Antonio, P.; Quartucci, G.; Capozzi, V. Characterization of human cells exposed to deltamethrin by means of Raman microspectroscopy and atomic force microscopy. Vib. Spectrosc. 2011, 57, 55–60. [Google Scholar] [CrossRef]
- Kawamura, K.; Tsujimoto, Y.; Rabenarivo, M.; Asai, H.; Andriamananjara, A.; Rakotoson, T. Vis-NIR Spectroscopy and PLS Regression with Waveband Selection for Estimating the Total C and N of Paddy Soils in Madagascar. Remote Sens. 2017, 9, 1081. [Google Scholar] [CrossRef]
- Vivótruyols, G.; Schoenmakers, P.J. Automatic selection of optimal savitzky-golay smoothing. Anal. Chem. 2006, 78, 4598–4608. [Google Scholar] [CrossRef] [PubMed]
- Fearn, T.; Riccioli, C.; Garrido-Varo, A.; Guerrero-Ginel, J.E. On the geometry of SNV and MSC. Chemom. Intell. Lab. Syst. 2009, 96, 22–26. [Google Scholar] [CrossRef]
- Nie, P.; Tao, D.; Yong, H.; Qu, F. Detection of soil nitrogen using near infrared sensors based on soil pretreatment and algorithms. Sensors 2017, 17, 1102. [Google Scholar] [CrossRef] [PubMed]
- Dettman, J.W.; Balise, P.L. Mathematical methods in physics and engineering. Phys. Today 1965, 16, 78–80. [Google Scholar] [CrossRef]
- Zou, X.; Zhao, J.; Li, Y. Selection of the efficient wavelength regions in FT-NIR spectroscopy for determination of SSC of ‘Fuji’ apple based on BIPLS and FIPLS models. Vib. Spectrosc. 2007, 44, 220–227. [Google Scholar] [CrossRef]
- Xiao, S.; He, Y.; Dong, T.; Nie, P. Spectral analysis and sensitive waveband determination based on nitrogen detection of different soil types using near infrared sensors. Sensors 2018, 18, 523. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Behera, S.K.; Pratihar, S.K.; Bhattacharyya, S. Low temperature synthesis of microwave dielectric laalo nanoparticles: Effect of chloride on phase evolution and morphology. Ceram. Int. 2004, 30, 1231–1235. [Google Scholar] [CrossRef]
- Su, Q.; Ma, X.; Dong, J.; Jiang, C.; Qian, W. A reproducible SERS substrate based on electrostatically assisted aptes-functionalized surface-assembly of gold nanostars. ACS Appl. Mater. Interfaces 2011, 3, 1873–1879. [Google Scholar] [CrossRef] [PubMed]
- Ipatov, A.; Cordova, F.; Doriol, L.J.; Casida, M.E. Excited-state spin-contamination in time-dependent density-functional theory for molecules with open-shell ground states. J. Mol. Struct. Theochem. 2009, 914, 60–73. [Google Scholar] [CrossRef]
- Kang, X.; Huang, J.; Shi, Y.Q.; Lin, Y. A DWT-DFT composite watermarking scheme robust to both affine transform and jpeg compression. IEEE Trans. Circ. Syst. Video Technol. 2003, 13, 776–786. [Google Scholar] [CrossRef]
- Ropo, M.; Kokko, K.; Vitos, L. Proving the perdew-burke-ernzerhof density functional designed for metallic bulk and surface systems. Physics 2007, 77, 998–1002. [Google Scholar]
- Kneipp, J.; Kneipp, H.; Kneipp, K. SERS—A single-molecule and nanoscale tool for bioanalytics. Chem. Soc. Rev. 2008, 37, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Dong, T.; Nie, P.; Qu, F.; He, Y.; Chu, B. Rapid determination of thiabendazole pesticides in rape by surface enhanced Raman spectroscopy. Sensors 2018, 18, 1082. [Google Scholar] [CrossRef] [PubMed]
- Parsons, H.M.; Ekman, D.R.; Collette, T.W.; Viant, M.R. Spectral relative standard deviation: A practical benchmark in metabolomics. Analyst 2009, 134, 478–485. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
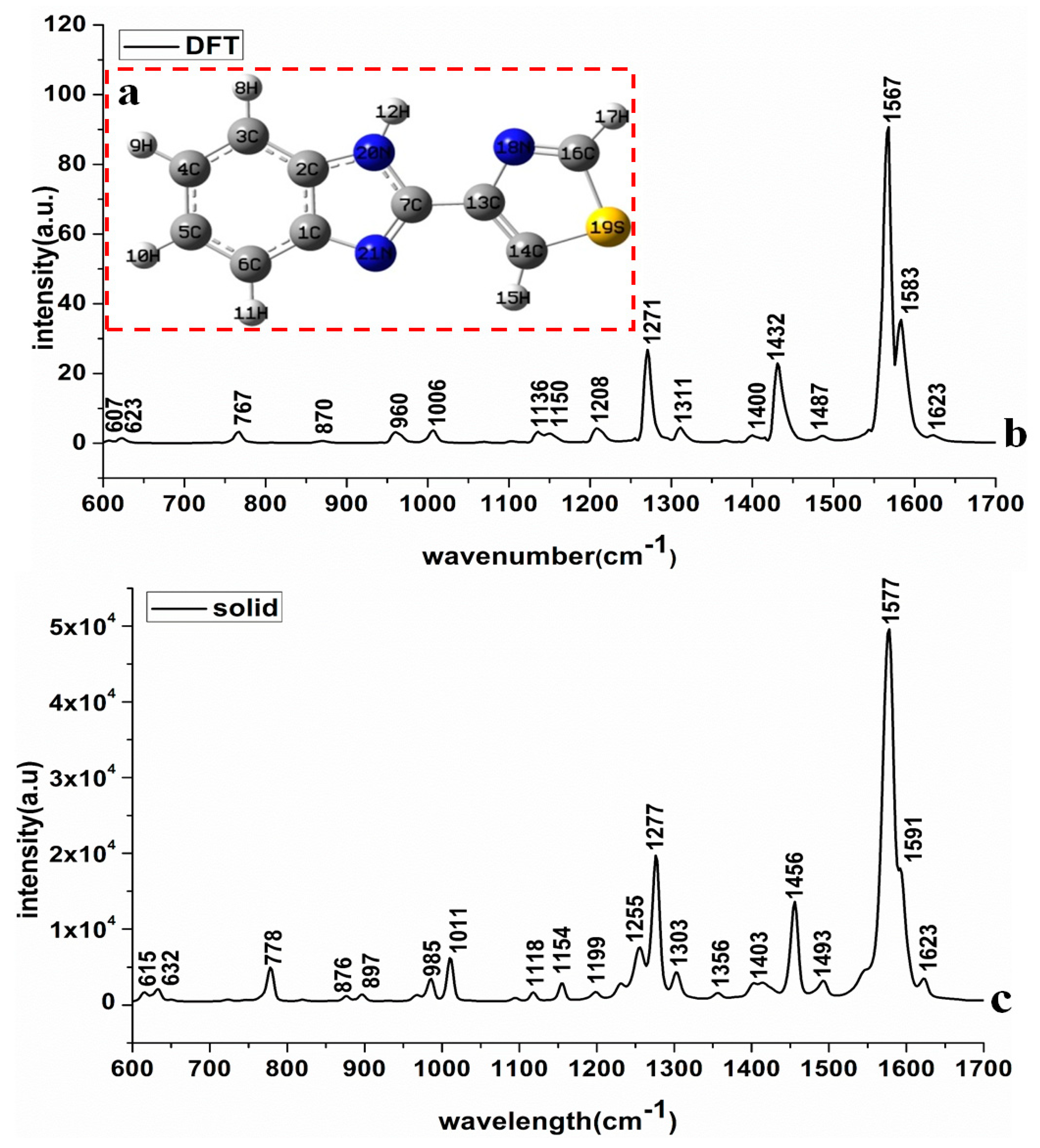





| Calculation (cm−1) | Solid (cm−1) | SERS-Au (cm−1) | Assignments |
|---|---|---|---|
| 607 (vs) | 615 (w) | - | δ(C-C-C)opp + δ(S-C-N)ip |
| 623 (vs) | 632 (w) | 626 (w) | δ(C-C-C)opp + δ(S-C-N)ip |
| 767 (w) | 778 (m) | 784 (m) | δ(C-H)oop |
| 870 (vs) | 876 (w) | 850 (m) | δ (C-C-C)opp + δ(C-H)opp |
| - | 897 (w) | 903 (m) | δ (C-C-C)opp + δ(C-H)opp |
| 960 (w) | 985 (m) | - | υ(C-S) |
| 1006 (m) | 1011 (m) | 1008 (s) | δ(C-H)ip |
| 1135 (w) | 1118 (m) | 1115 (w) | δ(C-H)ip |
| 1150 (m) | 1154 (m) | 1146 (w) | δ(C-H)ip |
| 1208 (w) | 1199 (m) | 1198 (w) | υ ring |
| - | - | 1220 (w) | υ ring |
| - | 1255 (m) | - | υ ring + δ(C-H)ip |
| 1271 (s) | 1277 (s) | 1270 (m) | υ ring + δ(C-H)ip |
| 1311 (w) | 1303 (w) | 1328 (m) | δ(C-H)ip |
| 1400 (w) | 1403 (w) | 1408 (m) | υ(C=C) |
| 1432 (s) | 1456 (s) | - | υ(C=N) |
| 1487 (w) | 1493 (w) | 1496 (w) | υ(C=C) + δ(N-H)ip |
| 1567 (vs) | 1577 (vs) | 1567 (s) | υ(C=N) |
| 1583 (s) | 1591 (s) | 1595 (m) | υ(C=N) |
| 1623 (w) | 1623 (w) | 1620 (w) | υ(C=N) |
| Types | Number | Min (nm) | Max (nm) | Average (nm) | Standard Deviation (nm) |
|---|---|---|---|---|---|
| AuNPs | 41 | 16.7 | 36.7 | 27.8 | 5.6 |
| Model | Sample | Predicted Value (mg/L) | RSD (%) | Recovery (%) | ||||
|---|---|---|---|---|---|---|---|---|
| True Value (mg/L) | Number | Min | Max | Mean | Min | Max | ||
| y = 672.26x + 5748.4 (784 cm−1) | 2 | 3 | 1.91 | 2.33 | 2.12 | 8.02 | 95.90 | 116.65 |
| 6 | 3 | 6.00 | 6.18 | 6.07 | 1.36 | 100.46 | 103.16 | |
| y = 1155.4x + 8740.2 (1008 cm−1) | 2 | 3 | 1.91 | 2.22 | 2.07 | 5.96 | 95.73 | 110.83 |
| 6 | 3 | 5.92 | 6.28 | 6.08 | 2.41 | 98.66 | 104.59 | |
| y = 784.97x + 6935.2 (1270 cm−1) | 2 | 3 | 1.77 | 2.14 | 1.92 | 8.23 | 88.25 | 106.78 |
| 6 | 3 | 5.19 | 5.82 | 5.40 | 5.45 | 86.64 | 96.97 | |
| y = 535.17x + 6798 (1328 cm−1) | 2 | 3 | 1.87 | 2.27 | 2.11 | 8.34 | 93.25 | 113.33 |
| 6 | 3 | 5.01 | 5.67 | 5.26 | 5.58 | 83.46 | 94.57 | |
| y = 385.33x + 6277.3 (1406 cm−1) | 2 | 3 | 1.73 | 1.89 | 1.83 | 3.84 | 86.62 | 94.65 |
| 6 | 3 | 4.66 | 4.76 | 4.76 | 3.84 | 77.66 | 79.26 | |
| y = 607.13x + 6248.3 (1564 cm−1) | 2 | 3 | 1.44 | 1.71 | 1.56 | 7.39 | 71.87 | 85.57 |
| 6 | 3 | 4.99 | 6.19 | 5.39 | 10.45 | 83.13 | 103.19 | |
| Methods | Pre-Processing Method | Calibration | Prediction | |||
|---|---|---|---|---|---|---|
| RC2 | RMSEC (mg/L) | RP2 | RMSEP (mg/L) | RPD | ||
| PLS | Original | 0.9600 | 0.607 | 0.9549 | 0.694 | 4.79 |
| S-G | 0.9580 | 0.622 | 0.9528 | 0.691 | 4.65 | |
| MSC | 0.9592 | 0.647 | 0.9683 | 0.678 | 4.62 | |
| SNV | 0.9493 | 0.711 | 0.9769 | 0.556 | 5.97 | |
| biPLS | Original | 0.9518 | 0.698 | 0.9746 | 0.523 | 6.13 |
| S-G | 0.9532 | 0.684 | 0.9657 | 0.605 | 5.39 | |
| MSC | 0.9633 | 0.600 | 0.9580 | 0.683 | 5.01 | |
| SNV | 0.9415 | 0.741 | 0.9754 | 0.561 | 6.29 | |
| Sample | Ture Value (mg/L) | Predicted Value (mg/L) | RSD (%) | Recovery (%) |
|---|---|---|---|---|
| 1 | 1 | 1.12 | 12 | 112 |
| 2 | 2 | 2.119 | 5.9 | 105.95 |
| 3 | 2.5 | 2.253 | 9.8 | 90.12 |
| 4 | 5 | 4.46 | 10.8 | 89.2 |
| 5 | 7.5 | 7.57 | 0.93 | 100.93 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, P.; Dong, T.; Xiao, S.; Lin, L.; He, Y.; Qu, F. Quantitative Determination of Thiabendazole in Soil Extracts by Surface-Enhanced Raman Spectroscopy. Molecules 2018, 23, 1949. https://doi.org/10.3390/molecules23081949
Nie P, Dong T, Xiao S, Lin L, He Y, Qu F. Quantitative Determination of Thiabendazole in Soil Extracts by Surface-Enhanced Raman Spectroscopy. Molecules. 2018; 23(8):1949. https://doi.org/10.3390/molecules23081949
Chicago/Turabian StyleNie, Pengcheng, Tao Dong, Shupei Xiao, Lei Lin, Yong He, and Fangfang Qu. 2018. "Quantitative Determination of Thiabendazole in Soil Extracts by Surface-Enhanced Raman Spectroscopy" Molecules 23, no. 8: 1949. https://doi.org/10.3390/molecules23081949
APA StyleNie, P., Dong, T., Xiao, S., Lin, L., He, Y., & Qu, F. (2018). Quantitative Determination of Thiabendazole in Soil Extracts by Surface-Enhanced Raman Spectroscopy. Molecules, 23(8), 1949. https://doi.org/10.3390/molecules23081949






