‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins
Abstract
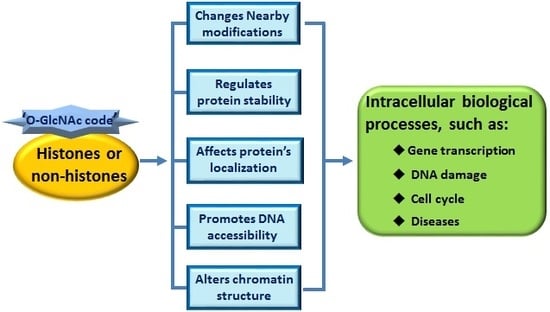
:1. Introduction
2. Add and Remove: Regulation of ‘O-GlcNAc Code’ in Cells
3. ‘O-GlcNAc Code’ Provides a Recognition Platform to Initiate Subsequent Functions
3.1. ‘O-GlcNAc Code’ on Histones
3.2. OGT Assembled in Different Protein Complexes to Coordinate Specific Functions
3.3. Function Switching between O-GlcNAcylation and Phosphorylation
3.4. ‘O-GlcNAc Code’ on Non-Histone Proteins Targets Specific Biological Functions
4. ‘O-GlcNAc Code’ Mediated Diseases
4.1. ‘O-GlcNAc Code’ in Cancer
4.2. ‘O-GlcNAc Code’ in Diabetes
4.3. ‘O-GlcNAc Code’ in Alzheimer’s Disease
4.4. ‘O-GlcNAc Code’ in Cardiovascular Diseases
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Butkinaree, C.; Park, K.; Hart, G.W. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 2010, 1800, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef] [PubMed]
- Haltiwanger, R.S.; Blomberg, M.A.; Hart, G.W. Glycosylation of Nuclear and Cytoplasmic Proteins Purification and characterization of a uridine diphospho-N-acetylglucosamine: polypeptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992, 267, 9005–9013. [Google Scholar] [PubMed]
- Lefebvre, T.; Drougat, L.; Olivier-Van Stichelen, S.; Michalski, J.C.; Vercoutter-Edouart, A.S. Antibodies and activity measurements for the detection of O-GlcNAc transferase and assay of itssubstrate, UDP-GlcNAc. Methods Mol. Biol. 2013, 1022, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Wells, L.; Gao, Y.; Mahoney, J.A.; Vosseller, K.; Chen, C.; Rosen, A.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: Further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002, 277, 1755–1761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tan, E.P.; VandenHull, N.J.; Peterson, K.R.; Slawson, C. O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. (Lausanne) 2014, 5, 206. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.A.; Hanover, J.A. O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 2014, 289, 34440–34448. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.A. O-GlcNAcylation at Promoters, Nutrient Sensors, and Transcriptional Regulation. Biochim. Biophys. Acta 2013, 1829, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Hardivillé, S.; Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell. MeTable 2014, 20, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Kim, J.E.; Nam, H.W.; Ju, J.W.; Kim, H.S.; Kim, Y.S.; Cho, J.W. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006, 8, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Qiu, Z.; Zhang, S.; Fan, X.; Cai, X.; Xu, B.; Li, X.; Zhou, J.; Zhang, X.; Chu, Y.; et al. Elevated O-GlcNAcylation promotes gastric cancer cells proliferation by modulating cell cycle related proteins and ERK 1/2 signaling. Oncotarget 2016, 7, 61390–61402. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Zhu, W.; Anderson, R.A.; Leber, B.; Andrews, D.W. Multiple post-translational modifications regulate E-cadherin transport during apoptosis. J. Cell. Sci. 2012, 125, 2615–2625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.P.; Zhang, K.; Wu, J.; Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015, 356, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Slawson, C.; Lakshmanan, T.; Knapp, S.; Hart, G.W. A Mitotic GlcNAcylation/Phosphorylation Signaling Complex Alters the Posttranslational State of the Cytoskeletal Protein Vimentin. Mol. Biol. Cell 2008, 19, 4130–4140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisi, V.; Miseta, A.; Nagy, T. The Role of Stress-Induced O-GlcNAc Protein Modification in the Regulation of Membrane Transport. Oxid. Med. Cell. Longev. 2017, 2017, 1308692. [Google Scholar] [CrossRef] [PubMed]
- Wells, L.; Vosseller, K.; Hart, G.W. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 2003, 60, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhao, L.; Feng, Z.; Yu, C.; Ding, J.; Wang, L.; Wang, F.; Liu, D.; Zhu, H.; Xing, F.; et al. O-Linked N-acetylglucosamine transferase 1 regulates global histone H4 acetylation via stabilization of the nonspecific lethal protein NSL3. J. Biol. Chem. 2017, 292, 10014–10025. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, P.; Yan, H.; Sun, H.; Liu, X.; Zhou, F.; Li, L.; Chen, Y.; Muthana, M.M.; Chen, X.; et al. Substrate Specificity Provides Insights into the Sugar Donor Recognition Mechanism of O-GlcNAc Transferase (OGT). PLoS ONE 2013, 8, e63452. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.C.; Yu, S.H.; Li, B.; Zegzouti, H.; Kohler, J.J. Enhanced Transfer of a Photocross-linking N-Acetylglucosamine (GlcNAc) Analog by an O-GlcNAc Transferase Mutant with Converted Substrate Specificity. J. Biol. Chem. 2015, 290, 22638–22648. [Google Scholar] [CrossRef] [PubMed]
- Hanover, J.A.; Yu, S.; Lubas, W.B.; Shin, S.H.; Ragano-Caracciola, M.; Kochran, J.; Love, D.C. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 2003, 409, 287–297. [Google Scholar] [CrossRef]
- Lazarus, M.B.; Nam, Y.; Jiang, J.; Sliz, P.; Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Kapuria, V.; Röhrig, U.F.; Bhuiyan, T.; Borodkin, V.S.; van Aalten, D.M.; Zoete, V.; Herr, W. Proteolysis of HCF-1 by Ser/Thr glycosylation-incompetent O-GlcNAc transferase: UDP-GlcNAc complexes. Genes Dev. 2016, 30, 960–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riu, I.H.; Shin, I.S.; Do, S.I. Sp1 modulates ncOGT activity to alter target recognition and enhanced thermotolerance in E. coli. Biochem. Biophys. Res. Commun. 2008, 372, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Sacoman, J.L.; Dagda, R.Y.; Burnham-Marusich, A.R.; Dagda, R.K.; Berninsone, P.M. Mitochondrial O-GlcNAc transferase (mOGT) regulates mitochondrial structure, function and survival in HeLa cells. J. Biol. Chem. 2017, 292, 4499–4518. [Google Scholar] [CrossRef] [PubMed]
- Trapannone, R.; Mariappa, D.; Ferenbach, A.T.; van Aalten, D.M. Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochem. J. 2016, 473, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Kreppel, L.K.; Hart, G.W. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 1999, 274, 32015–32022. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.P.; Hart, G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 2003, 278, 24608–24616. [Google Scholar] [CrossRef] [PubMed]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Méndez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013, 32, 645–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burén, S.; Gomes, A.L.; Teijeiro, A.; Fawal, M.A.; Yilmaz, M.; Tummala, K.S.; Perez, M.; Rodriguez-Justo, M.; Campos-Olivas, R.; Megías, D.; et al. Regulation of OGT by URI in Response to Glucose Confers c-MYC-Dependent Survival Mechanisms. Cancer Cell 2016, 30, 290–307. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Swanson, S.K.; Cole, M.D.; Choi, S.H.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 2010, 285, 4268–4272. [Google Scholar] [CrossRef] [PubMed]
- Comtesse, N.; Maldener, E.; Meese, E. Identification of a nuclear variant of MGEA5: A cytoplasmic hyaluronidase and a beta-Nacetylglucosaminidase. Biochem. Biophys. Res. Commun. 2001, 283, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Toleman, C.; Paterson, A.J.; Whisenhunt, T.R.; Kudlow, J.E. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 2004, 279, 53665–53673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Roche, K.; Nasheuer, H.P.; Lowndes, N.F. Modification of Histones by Sugar-N-Acetylglucosamine (GlcNAc) Occurs on Multiple Residues, Including Histone H3 Serine 10, and Is Cell Cycle-regulated. J. Biol. Chem. 2011, 286, 37483–37495. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, R.; Hashiba, W.; Sekine, H.; Yokoyama, A.; Chikanishi, T.; Ito, S.; Imai, Y.; Kim, J.; He, H.H.; Igarashi, K.; et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 2011, 480, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, X. OGT restrains the expansion of DNA damage signaling. Nucleic Acids Res. 2016, 44, 9266–9278. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Z.; Li, L.; Gong, W.; Lazenby, A.J.; Swanson, B.J.; Herring, L.E.; Asara, J.M.; Singer, J.D.; Wen, H. Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J. Exp. Med. 2017, 214, 1093–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambetta, M.C.; Müller, J. A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma 2015, 124, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, K.; Wang, Z.; Hart, G.W. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. USA 2010, 107, 19915–19920. [Google Scholar] [CrossRef] [PubMed]
- Lercher, L.; Raj, R.; Patel, N.A.; Price, J.; Mohammed, S.; Robinson, C.V.; Schofield, C.J.; Davis, B.G. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun. 2015, 6, 7978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, K.; Kato, A.; Kobayashi, J.; Yanagihara, H.; Sakamoto, S.; Oliveira, D.V.; Shimada, M.; Tauchi, H.; Suzuki, H.; Tashiro, S.; et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Mol. Cell 2011, 41, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Hirosawa, M.; Hayakawa, K.; Yoneda, C.; Arai, D.; Shiota, H.; Suzuki, T.; Tanaka, S.; Dohmae, N.; Shiota, K. Novel O-GlcNAcylation on Ser (40) of canonical H2A isoforms specific to viviparity. Sci. Rep. 2016, 6, 31785. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.T.; Kim, H.; Lu, W.; He, Q.; Liu, D.; Goodell, M.A.; Wan, M.; Songyang, Z. Ten-Eleven Translocation 1 (Tet1) Is Regulated by O-Linked N-Acetylglucosamine Transferase (Ogt) for Target Gene Repression in Mouse Embryonic Stem Cells. J. Biol. Chem. 2013, 288, 20776–20784. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Christensen, J.; Helin, K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011, 13, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, Y.; Bian, C.; Fujiki, R.; Yu, X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 2013, 493, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Capotosti, F.; Guernier, S.; Lammers, F.; Waridel, P.; Cai, Y.; Jin, J.; Conaway, J.W.; Conaway, R.C.; Herr, W. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 2011, 144, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.B.; Han, X.; Li, M.D.; Singh, J.P.; Qian, K.; Azarhoush, S.; Zhao, L.; Bennett, A.M.; Samuel, V.T.; Wu, J.; et al. O-GlcNAc Transferase/Host Cell Factor C1 Complex Regulates Gluconeogenesis by Modulating PGC-1α Stability. Cell. MeTable 2012, 16, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Mazars, R.; Gonzalez-de-Peredo, A.; Cayrol, C.; Lavigne, A.C.; Vogel, J.L.; Ortega, N.; Lacroix, C.; Gautier, V.; Huet, G.; Ray, A.; et al. The THAP-Zinc Finger Protein THAP1 Associates with Coactivator HCF-1 and O-GlcNAc Transferase, a link between DYT6 and DYT3 dystonias. J. Biol. Chem. 2010, 285, 13364–13371. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jiang, W.; Zhou, P.; Liu, L.; Wan, X.; Yuan, X.; Wang, X.; Chen, M.; Chen, J.; Yang, J.; et al. Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PLoS ONE. 2015, 10, e0145023. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Shimoji, S.; Hart, G.W. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010, 584, 2526–2538. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, K.; Hart, G.W. O-GlcNAc Transferase Regulates Mitotic Chromatin Dynamics. J. Biol. Chem. 2010, 285, 34460–34468. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.J.; Nguyen, B.L.; Bridger, R.; Medrano, E.E.; Wells, L.; Pan, S.; Sifers, R.N. β-N-Acetylglucosamine (O-GlcNAc) Is a Novel Regulator of Mitosis-specific Phosphorylations on Histone H3. J. Biol. Chem. 2012, 287, 12195–12203. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Earnshaw, W.C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003, 4, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Tao, T.; Zhang, D.; Liu, X.; Qiu, H.; Han, L.; Xu, Z.; Xiao, Y.; Cheng, C.; Shen, A. O-GlcNAcylation of histone deacetylase-1 in hepatocellular carcinoma promotes cancer progression. Glycobiology 2016, 26, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Iitaka, C.; Miyazaki, K.; Akaike, T.; Ishida, N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J. Biol. Chem. 2005, 280, 29397–29402. [Google Scholar] [CrossRef] [PubMed]
- Kaasik, K.; Kivimäe, S.; Allen, J.J.; Chalkley, R.J.; Huang, Y.; Baer, K.; Kissel, H.; Burlingame, A.L.; Shokat, K.M.; Ptáček, L.J.; et al. Glucose Sensor O-GlcNAcylation Coordinates with Phosphorylation to Regulate Circadian Clock. Cell MeTable 2013, 17, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Smet-Nocca, C.; Broncel, M.; Wieruszeski, J.M.; Tokarski, C.; Hanoulle, X.; Leroy, A.; Landrieu, I.; Rolando, C.; Lippens, G.; Hackenberger, C.P. Identification of O-GlcNAc sites within peptides of the Tau protein and their impact on phosphorylation. Mol. Biosyst. 2011, 7, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.; Daou, S.; Zamorano, N.; Iannantuono, N.V.; Hammond-Martel, I.; Mashtalir, N.; Bonneil, E.; Wurtele, H.; Thibault, P.; Affar el, B. Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics 2015, 10, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Vosseller, K.; Sakabe, K.; Wells, L.; Hart, G.W. Diverse regulation of protein function by O-GlcNAc: A nuclear and cytoplasmic carbohydrate post-translational modification. Curr. Opin. Chem. Biol. 2002, 6, 851–857. [Google Scholar] [CrossRef]
- Howerton, C.L.; Morgan, C.P.; Fischer, D.B.; Bale, T.L. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. USA 2013, 110, 5169–5174. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.S.; Lo, P.W.; Yeh, Y.H.; Hsu, P.H.; Peng, S.H.; Teng, Y.C.; Kang, M.L.; Wong, C.H.; Juan, L.J. O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. USA 2014, 111, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Charoensuksai, P.; Kuhn, P.; Wang, L.; Sherer, N.; Xu, W. O-GlcNAcylation of co-activator-associated arginine methyltransferase 1 regulates its protein substrate specificity. Biochem. J. 2015, 466, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Schurter, B.T.; Koh, S.S.; Chen, D.; Bunick, G.J.; Harp, J.M.; Hanson, B.L.; Henschen-Edman, A.; Mackay, D.R.; Stallcup, M.R.; Aswad, D.W. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 2001, 40, 5747–5756. [Google Scholar] [CrossRef]
- Sebastian, S.; Sreenivas, P.; Sambasivan, R.; Cheedipudi, S.; Kandalla, P.; Pavlath, G.K.; Dhawan, J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 4719–4724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wong, J.; Klinger, M.; Tran, M.T.; Shannon, K.M.; Killeen, N. Mll5 contributes to hematopoietic stem cell fitness and homeostasis. Blood 2009, 113, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, M.D.; Marmorstein, R. A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function. J. Mol. Biol. 2017, 429, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Wang, H.; Li, J.; Holec, S.; Berger, F. The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol. Open 2014, 3, 794–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.S.; Zhang, Z. O-linked N-acetylglucosamine transferase (OGT) interacts with the histone chaperone HIRA complex and regulates nucleosome assembly and cellular senescence. Proc. Natl. Acad. Sci. USA. 2016, 113, E3213–E3220. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, H.; Li, J.; Wang, Y.; Ding, Y.H.; Shen, H.; Yang, Y.; Sun, C.; Huang, M.; Tu, Y.; et al. Polη O-GlcNAcylation governs genome integrity during translesion DNA synthesis. Nat. Commun. 2017, 8, 1941. [Google Scholar] [CrossRef] [PubMed]
- Livneh, Z.; Ziv, O.; Shachar, S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle 2010, 9, 729–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Gu, J.H.; Dai, C.L.; Gu, J.; Jin, X.; Sun, J.; Iqbal, K.; Liu, F.; Gong, C.X. O-GlcNAcylation regulates ischemia-induced neuronal apoptosis through AKT signaling. Sci. Rep. 2015, 5, 14500. [Google Scholar] [CrossRef] [PubMed]
- Brembeck, F.H.; Rosário, M.; Birchmeier, W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 2006, 16, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Olivier-Van Stichelen, S.; Dehennaut, V.; Buzy, A.; Zachayus, J.L.; Guinez, C.; Mir, A.M.; El Yazidi-Belkoura, I.; Copin, M.C.; Boureme, D.; Loyaux, D.; et al. O-GlcNAcylation stabilizes β-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 2014, 28, 3325–3338. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, L.; Zhou, C.; Lin, N.; Liu, A. Sirt 1 activator inhibits the AGE-induced apoptosis and p53 acetylation in human vascular endothelial cells. J. Toxicol. Sci. 2015, 40, 615–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, N.; Zhu, X.; He, W.; Zhao, S.; Zhao, W.; Zhu, C. Resveratrol inhibits the hydrogen dioxide-induced apoptosis via Sirt 1 activation in osteoblast cells. Biosci. Biotechnol. Biochem. 2015, 79, 1779–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.; Gu, Y.; Shan, H.; Mi, W.; Sun, J.; Shi, M.; Zhang, X.; Lu, X.; Han, F.; Gong, Q.; et al. O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat. Commun. 2017, 8, 1491. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wells, L.; Comer, F.I.; Parker, G.J.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 2001, 276, 9838–9845. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. (September). “p53 isoforms can regulate p53 transcriptional activity”. Gene Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Gu, W. Dual roles of MDM2 in the regulation of p53: Ubiquitination dependent and ubiquitination independent mechanisms of MDM2 repression of p53 activity. Genes Cancer 2012, 3, 240–248. [Google Scholar] [CrossRef] [PubMed]
- De Queiroz, R.M.; Madan, R.; Chien, J.; Dias, W.B.; Slawson, C. Changes in O-Linked N-Acetylglucosamine (O-GlcNAc) Homeostasis Activate the p53 Pathway in Ovarian Cancer Cells. J. Biol. Chem. 2016, 291, 18897–18914. [Google Scholar] [CrossRef] [PubMed]
- Parrales, A.; Iwakuma, T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front. Oncol. 2015, 5, 288. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, C.M.; Lu, T.Y.; Bacigalupa, Z.A.; Katsetos, C.D.; Sinclair, D.A.; Reginato, M.J. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene 2017, 36, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Itkonen, H.M.; Minner, S.; Guldvik, I.J.; Sandmann, M.J.; Tsourlakis, M.C.; Berge, V.; Svindland, A.; Schlomm, T.; Mills, I.G. O-GlcNAc Transferase Integrates Metabolic Pathways to Regulate the Stability of c-MYC in Human Prostate Cancer Cells. Cancer Res. 2013, 73, 5277–5287. [Google Scholar] [CrossRef] [PubMed]
- Sodi, V.L.; Khaku, S.; Krutilina, R.; Schwab, L.P.; Vocadlo, D.J.; Seagroves, T.N.; Reginato, M.J. mTOR/MYC Axis Regulates O-GlcNAc Transferase (OGT) Expression and O-GlcNAcylation in Breast Cancer. Mol. Can. Res. 2015, 13, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Hou, K.; Li, Z.; Li, L.; Yu, H.; Liu, Y.; Li, Y.; Yin, Z. Silencing β-linked N-acetylglucosamine transferase induces apoptosis in human gastric cancer cells through PUMA and caspase-3 pathways. Oncol. Rep. 2015, 34, 3140–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phoomak, C.; Vaeteewoottacharn, K.; Sawanyawisuth, K.; Seubwai, W.; Wongkham, C.; Silsirivanit, A.; Wongkham, S. Mechanistic insights of O-GlcNAcylation that promote progression of cholangiocarcinoma cells via nuclear translocation of NF-κB. Sci. Rep. 2016, 6, 27853. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Hart, G.W. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteom. 2013, 10, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, T.; Dehennaut, V.; Guinez, C.; Olivier, S.; Drougat, L.; Mir, A.M.; Mortuaire, M.; Vercoutter-Edouart, A.S.; Michalski, J.C. Dysregulation of the nutrient/stress sensor O-GlcNAcylation is involved in the etiology of cardiovascular disorders, type-2 diabetes and Alzheimer’s disease. Biochim. Biophys. Acta 2009, 1800, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Ryu, J.; Lee, W. O-GlcNAc modification on IRS-1 and Akt2 by PUGNAc inhibits their phosphorylation and induces insulin resistance in rat primary adipocytes. Exp. Mol. Med. 2005, 37, 220–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Huang, X.; Sun, D.; Xin, X.; Pan, Q.; Peng, S.; Liang, Z.; Luo, C.; Yang, Y.; Jiang, H.; et al. Extensive Crosstalk between O-GlcNAcylation and Phosphorylation Regulates Akt Signaling. PLoS ONE 2012, 7, e37427. [Google Scholar] [CrossRef] [PubMed]
- Andrali, S.S.; Qian, Q.; Ozcan, S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 2007, 282, 15589–15596. [Google Scholar] [CrossRef] [PubMed]
- Kebede, M.; Ferdaoussi, M.; Mancini, A.; Alquier, T.; Kulkarni, R.N.; Walker, M.D.; Poitout, V. Glucose activates free fatty acid receptor 1 gene transcription via phosphatidylinositol-3-kinase-dependent O-GlcNAcylation of pancreas-duodenum homeobox-1. Proc. Natl. Acad. Sci. USA 2012, 109, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, K.; Wells, L. Multiple Tissue-specific Roles for the O-GlcNAc Posttranslational Modification in the Induction of and Complications Arising from Type II Diabetes. J. Biol. Chem. 2014, 289, 34466–34471. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Saudek, C.D.; Hart, G.W. Increased Expression of β-N-Acetylglucosaminidase in Erythrocytes from Individuals With Pre-diabetes and Diabetes. Diabetes. 2010, 59, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.; Iliffe, S. Alzheimer’s disease. BMJ 2009, 338, B158. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shan, X.; Yuzwa, S.A.; Vocadlo, D.J. The emerging link between O-GlcNAc and Alzheimer disease. J. Biol. Chem. 2014, 289, 34472–34481. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.C.; Jensen, E.H.; Rexach, J.E.; Vinters, H.V.; Hsieh-Wilson, L.C. Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc. Natl. Acad. Sci. USA 2016, 113, 15120–15125. [Google Scholar] [CrossRef] [PubMed]
- Lo, R.Y.; Hubbard, A.E.; Shaw, L.M.; Trojanowski, J.Q.; Petersen, R.C.; Aisen, P.S.; Weiner, M.W.; Jagust, W.J. Longitudinal change of biomarkers in cognitive decline. Arch. Neurol. 2011, 68, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Mosconi, L.; Pupi, A.; De Leon, M.J. Brain glucose hypometabolism and oxidative stress in preclinical Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2008, 1147, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Yuzwa, S.A.; Shan, X.; Macauley, M.S.; Clark, T.; Skorobogatko, Y.; Vosseller, K.; Vocadlo, D.J. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 2012, 8, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Yuzwa, S.A.; Yadav, A.K.; Skorobogatko, Y.; Clark, T.; Vosseller, K.; Vocadlo, D.J. Mapping O-GlcNAc modification sites on tau and generation of a site-specific O-GlcNAc tau antibody. Amino Acids 2011, 40, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Trinidad, J.C.; Barkan, D.T.; Gulledge, B.F.; Thalhammer, A.; Sali, A.; Schoepfer, R.; Burlingame, A.L. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteom. 2012, 11, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Rexach, J.E.; Clark, P.M.; Mason, D.E.; Neve, R.L.; Peters, E.C.; Hsieh-Wilson, L.C. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 2012, 8, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Francisco, H.; Kollins, K.; Varghis, N.; Vocadlo, D.; Vosseller, K.; Gallo, G. O-GlcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev. Neurobiol. 2009, 69, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Tallent, M.K.; Varghis, N.; Skorobogatko, Y.; Hernandez-Cuebas, L.; Whelan, K.; Vocadlo, D.J.; Vosseller, K. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J. Biol. Chem. 2009, 284, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Pekkurnaz, G.; Trinidad, J.C.; Wang, X.; Kong, D.; Schwarz, T.L. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 2014, 158, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Schwarz, T.L. O-GlcNAc Transferase Is Essential for Sensory Neuron Survival and Maintenance. J. Neurosci. 2017, 37, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Chatham, J.C.; Nöt, L.G.; Fülöp, N.; Marchase, R.B. Hexosamine biosynthesis and protein O-glycosylation the first line of defense against stress, ischemia and trauma. Shock 2008, 29, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Ngoh, G.A.; Hamid, T.; Prabhu, S.D.; Jones, S.P. O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death. Am. J. Physiol. Heart Circ. Physiol. 2009, 297, H1711–H1719. [Google Scholar] [CrossRef] [PubMed]
- Fulop, N.; Zhang, Z.; Marchase, R.B.; Chatham, J.C. Glucosamine cardioprotection in perfused rat hearts associated with increased o-linked n-acetylglucosamine protein modification and altered p38 activation. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H2227–H2236. [Google Scholar] [CrossRef] [PubMed]
- Watson, L.J.; Facundo, H.T.; Ngoh, G.A.; Ameen, M.; Brainard, R.E.; Lemma, K.M.; Long, B.W.; Prabhu, S.D.; Xuan, Y.T.; Jones, S.P. O-linked beta-n-acetylglucosamine transferase is indispensable in the failing heart. Proc. Natl. Acad. Sci. USA 2010, 107, 17797–17802. [Google Scholar] [CrossRef] [PubMed]
- Dassanayaka, S.; Jones, S.P. O-GlcNAc and the cardiovascular system. Pharmacol. Ther. 2014, 142, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Medford, HM.; Marsh, SA. The role of O-GlcNAc transferase in regulating the gene transcription of developing and failing hearts. Future Cardiol. 2014, 10, 801–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, L.J.; Long, B.W.; DeMartino, A.M.; Brittian, K.R.; Readnower, R.D.; Brainard, R.E.; Cummins, T.D.; Annamalai, L.; Hill, B.G.; Jones, S.P. Cardiomyocyte Ogt is essential for postnatal viability. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, 142–153. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds ...... are available from the authors. |


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Shah, J.A.; Cai, Y.; Jin, J. ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules 2018, 23, 1967. https://doi.org/10.3390/molecules23081967
Zhao L, Shah JA, Cai Y, Jin J. ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules. 2018; 23(8):1967. https://doi.org/10.3390/molecules23081967
Chicago/Turabian StyleZhao, Linhong, Junaid Ali Shah, Yong Cai, and Jingji Jin. 2018. "‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins" Molecules 23, no. 8: 1967. https://doi.org/10.3390/molecules23081967
APA StyleZhao, L., Shah, J. A., Cai, Y., & Jin, J. (2018). ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules, 23(8), 1967. https://doi.org/10.3390/molecules23081967