Prediction Methods of Herbal Compounds in Chinese Medicinal Herbs
Abstract
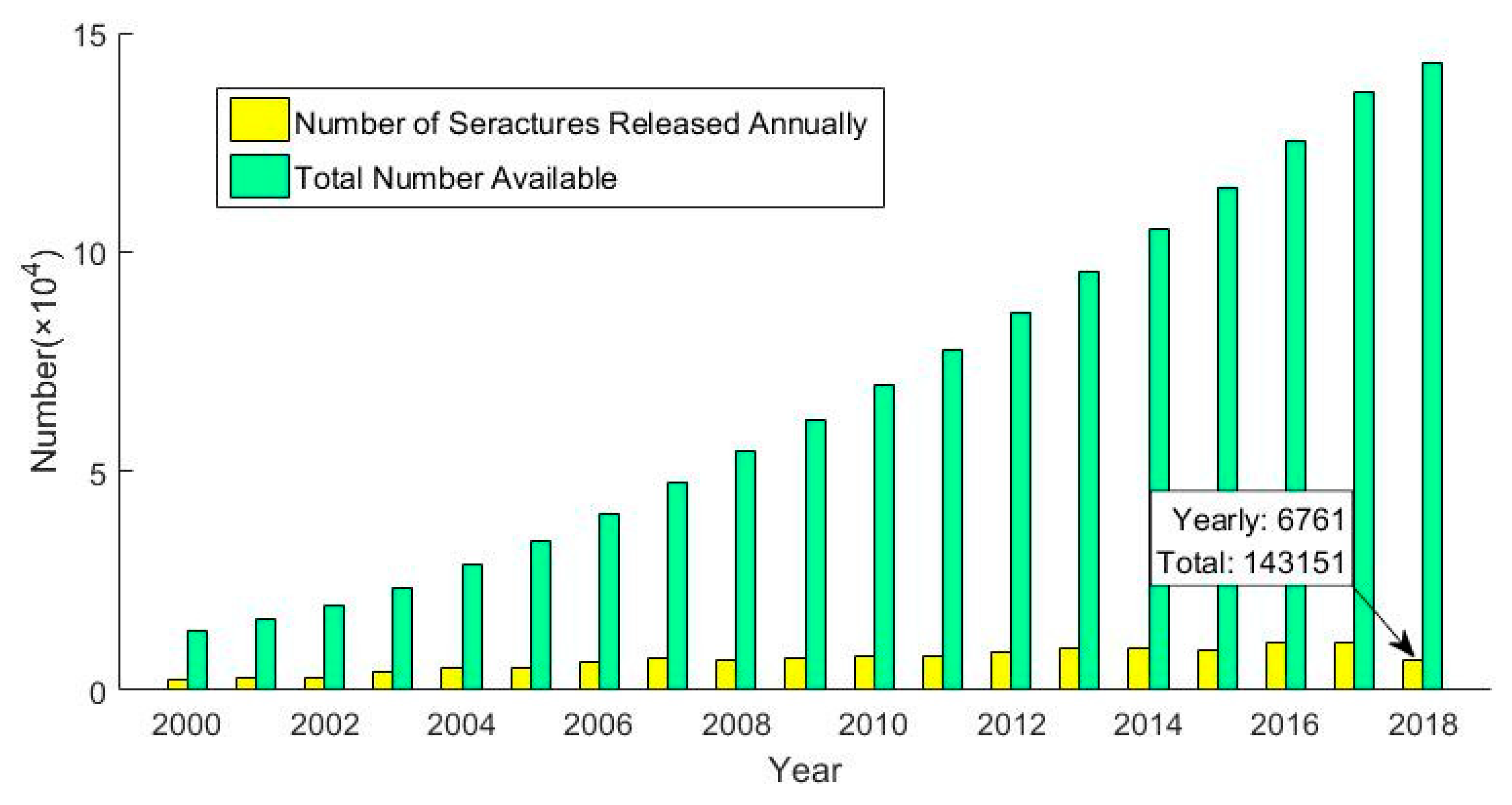
1. Introduction
2. Virtual Screening Technology
2.1. Target Selection
2.2. Ligand Selection
2.3. Molecular Docking
2.4. Virtual Screening Validation
2.5. Post-Testing and Processing
3. Introduce Other Prediction Methods of Herbal Compounds in Chinese Medicinal Herbs
3.1. Pharmacophore Model
3.2. Machine Learning Method
4. Conclusions
Funding
Conflicts of Interest
References
- Li, R.; Zhai, H.Q.; Tian, W.L.; Hou, J.R.; Jin, S.Y.; Wang, Y.Y. Comparative analysis between origin of cooked traditional Chinese medicine powder and modern formula granules. Zhongguo Zhong Yao Za Zhi 2016, 41, 965–969. [Google Scholar] [PubMed]
- Jorgensen, W.L. The many roles of computation in drug discovery. Science 2004, 303, 1813–1818. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Liu, D.; Li, L.; Wempe, M.F.; Guin, S.; Khanna, M.; Meier, J.; Hoffman, B.; Owens, C.; Wysoczynski, C.L.; et al. Discovery and characterization of small molecules that target the GTPaseRal. Nature 2014, 515, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Rui, Z. Processing Guide; Hainan Publishing House: Haikou, China, 2000. [Google Scholar]
- Chao, J.; Dai, Y.; Verpoorte, R.; Lam, W.; Cheng, Y.C.; Pao, L.H.; Zhang, W.; Chen, S. Major achievements of evidence-based traditional Chinese medicine in treating major diseases. Biochem. Pharmacol. 2017, 139, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Gao, Y.; Yang, J.M.; Meng, X.C. Overview of traditional Chinese medicine quality evaluation method based on overall research. Zhongguo Zhong Yao Za Zhi 2015, 40, 1027–1031. [Google Scholar] [PubMed]
- Hu, Y.N.; Ren, Y.L.; Cao, J.; Wang, M.; Wang, Y.; Qiao, Y.J. Predictive study on properties of traditional Chinese medicine components based on pharmacological effects. Zhongguo Zhong Yao Za Zhi 2014, 39, 2382–2385. [Google Scholar] [PubMed]
- Zhang, W.; Ji, L.; Chen, Y.; Tang, K.; Wang, H.; Zhu, R.; Jia, W.; Cao, Z.; Liu, Q. When drug discovery meets web search: Learning to rank for ligand-based virtual screening. J. Cheminform. 2015, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, P.B.; Ajay, M.K.; Nufail, M.; Gopakumar, G.; Jaleel, U.C. Gpurfscreen: A GPU based virtual screening tool using random forest classifier. J. Cheminform. 2016, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- SimhadriVsdna, N.; Muniappan, M.; Kannan, I.; Viswanathan, S. Phytochemical analysis and docking study of compounds present in a polyherbal preparation used in the treatment of dermatophytosis. Curr. Med. Mycol. 2017, 3, 6–14. [Google Scholar]
- Lee, H.S.; Im, W. G-LoSA: An efficient computational tool for local structure-centric biological studies and drug design. Protein Sci. 2016, 25, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; VijayaPrabhu, S.; Suryanarayanan, V.; Bhardwaj, R.; Singh, S.K.; Dubey, V.K. Molecular docking and structure-based virtual screening studies of potential drug target, CAAX prenyl proteases, of leishmaniadonovani. J. Biomol. Struct. Dyn. 2016, 34, 2367–2386. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv. Drug Deliv. Rev. 2016, 101, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y. Machine learning to improve the effectiveness of ANRS in predicting HIV drug resistance. Healthc. Inform. Res. 2017, 23, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.P.; Pokorny, A.; Teng, C.; Dear, R.; Epstein, R.J. Computational prediction of multidisciplinary team decision-making for adjuvant breast cancer drug therapies: A machine learning approach. BMC Cancer 2016, 16, 929. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.; Park, W.; Kang, T.W.; Cha, M.H.; Chun, J.M. Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. J. Ethnopharmacol. 2018, 221, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zeng, L.; Ge, J. Exploring the pharmacological mechanism of DanzhiXiaoyao powder on ER-positive breast cancer by a network pharmacology approach. Evid. Based Complement. Altern. Med. 2018, 2018, 5059743. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.L.; Fu, T.M.; Li, W.; Xu, X.L.; Sun, H.P. Discovery of new acetylcholinesterase inhibitors with small core structures through shape-based virtual screening. Bioorg. Med. Chem. Lett. 2015, 25, 3442–3446. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kang, H.; Liu, W.; Singhal, S.; Jiao, N.; Wang, Y.; Zhu, L.; Zhu, R. In silico design of novel proton-pump inhibitors with reduced adverse effects. Front. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Azad, I.; Nasibullah, M.; Khan, T.; Hassan, F.; Akhter, Y. Exploring the novel heterocyclic derivatives as lead molecules for design and development of potent anticancer agents. J. Mol. Graph. Model. 2018, 81, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Hofmann-Apitius, M.; Waldrich, O.; Ziegler, W. A robust framework for rapid deployment of a virtual screening laboratory. Stud. Health Technol. Inform. 2009, 147, 212–221. [Google Scholar] [PubMed]
- Huang, Y.X.; Zhao, J.; Song, Q.H.; Zheng, L.H.; Fan, C.; Liu, T.T.; Bao, Y.L.; Sun, L.G.; Zhang, L.B.; Li, Y.X. Virtual screening and experimental validation of novel histone deacetylase inhibitors. BMC Pharmacol. Toxicol. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Zu, S.; Chen, T.; Li, S. Global optimization-based inference of chemogenomic features from drug-target interactions. Bioinformatics 2015, 31, 2523–2529. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Xing, F.; Zeng, X.; Zou, Q. RicyerDB: A database for collecting rice yield-related genes with biological analysis. Int. J. Biol. Sci. 2018, 14, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A. Pay up or retract? Drug survey spurs conflict. Science 2017, 357, 1085–1086. [Google Scholar] [CrossRef] [PubMed]
- McKerrow, J.H.; Lipinski, C.A. The rule of five should not impede anti-parasitic drug development. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.; Ao, C.N.; Tou, K.I.; Huang, K.M.; Cheong, C.C.; Meng, L.R. Experimental and in silico analysis of cordycepin and its derivatives as endometrial cancer treatment. Oncol. Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Wu, X.; Qi, M.; Zhang, L.; Hu, H.; Zhao, Q.; Zhao, J.; Liu, H. Study on the mechanisms of active compounds in traditional chinese medicine for the treatment of influenza virus by virtual screening. Interdiscip. Sci. 2018, 10, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Onawole, A.T.; Kolapo, T.U.; Sulaiman, K.O.; Adegoke, R.O. Structure based virtual screening of the Ebola virus trimeric glycoprotein using consensus scoring. Comput. Biol. Chem. 2018, 72, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. The drug-maker’s guide to the galaxy. Nature 2017, 549, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Liu, S.; Zhang, G.; Bin, W.; Zhu, Y.; Frederick, D.T.; Hu, Y.; Zhong, W.; Randell, S.; Sadek, N.; et al. Pak signalling drives acquired drug resistance to MAPK inhibitors in BRAF-mutant melanomas. Nature 2017, 550, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Long, H.X.; Wang, M.; Fu, H.Y. Deep convolutional neural networks for predicting hydroxyproline in proteins. Curr. Bioinform. 2017, 12, 233–238. [Google Scholar] [CrossRef]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Khan, S.A.; Wennerberg, K.; Aittokallio, T. Global proteomics profiling improves drug sensitivity prediction: Results from a multi-omics, pan-cancer modeling approach. Bioinformatics 2018, 34, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, R.; Karunagaran, S.; Chandrabose, S.S.; Lee, K.W.; Meganathan, C. Pharmacophore modeling, virtual screening, molecular docking studies and density functional theory approaches to identify novel ketohexokinase (KHK) inhibitors. Biosystems 2015, 138, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, Y.; Xiong, X.; Liu, H.; Ran, T.; Xu, J.; Lu, S.; Xu, A.; Pan, J.; Qiao, X.; et al. Fragment virtual screening based on bayesian categorization for discovering novel vegfr-2 scaffolds. Mol. Diver. 2015, 19, 895–913. [Google Scholar] [CrossRef] [PubMed]
- Olayan, R.S.; Ashoor, H.; Bajic, V.B. DDR: Efficient computational method to predict drug-target interactions using graph mining and machine learning approaches. Bioinformatics 2018, 34, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, W.; Lin, H.; Wang, J.; Yang, Z.; Dumontier, M. Drug-drug interaction extraction via hierarchical RNNS on sequence and shortest dependency paths. Bioinformatics 2018, 34, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Rapakoulia, T.; Gao, X.; Huang, Y.; de Hoon, M.; Okada-Hatakeyama, M.; Suzuki, H.; Arner, E. Genome-scale regression analysis reveals a linear relationship for promoters and enhancers after combinatorial drug treatment. Bioinformatics 2017, 33, 3696–3700. [Google Scholar] [CrossRef] [PubMed]
- Dai, E.; Yang, F.; Wang, J.; Zhou, X.; Song, Q.; An, W.; Wang, L.; Jiang, W. NCDR: A comprehensive resource of non-coding RNAS involved in drug resistance. Bioinformatics 2017, 33, 4010–4011. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.F. Flexible receptor docking for drug discovery. Expert Opin. Drug Discov. 2015, 10, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Ferro, S.; Gitto, R.; Buemi, M.R.; Karamanou, S.; Stevaert, A.; Naesens, L.; De Luca, L. Identification of influenza PA-NTER endonuclease inhibitors using pharmacophore- and docking-based virtual screening. Bioorg. Med. Chem. 2018, 26, 4544–4550. [Google Scholar] [CrossRef] [PubMed]
- Spyrakis, F.; Benedetti, P.; Decherchi, S.; Rocchia, W.; Cavalli, A.; Alcaro, S.; Ortuso, F.; Baroni, M.; Cruciani, G. A pipeline to enhance ligand virtual screening: Integrating molecular dynamics and fingerprints for ligand and proteins. J. Chem. Inf. Model. 2015, 55, 2256–2274. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, T.; Selvam, C. Performance evaluation of structure based and ligand based virtual screening methods on ten selected anti-cancer targets. Bioorg. Med. Chem. Lett. 2015, 25, 4632–4636. [Google Scholar] [CrossRef] [PubMed]
- Temraz, M.G.; Elzahhar, P.A.; El-Din, A.B.A.; Bekhit, A.A.; Labib, H.F.; Belal, A.S.F. Anti-leishmanial click modifiable thiosemicarbazones: Design, synthesis, biological evaluation and in silico studies. Eur. J. Med. Chem. 2018, 151, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Ammad-Ud-Din, M.; Khan, S.A.; Wennerberg, K.; Aittokallio, T. Systematic identification of feature combinations for predicting drug response with Bayesian multi-view multi-task linear regression. Bioinformatics 2017, 33, i359–i368. [Google Scholar] [PubMed]
- Niinivehmas, S.P.; Salokas, K.; Latti, S.; Raunio, H.; Pentikainen, O.T. Ultrafast protein structure-based virtual screening with panther. J. Comput. Aided Mol. Des. 2015, 29, 989–1006. [Google Scholar] [PubMed]
- Cerqueira, N.M.; Gesto, D.; Oliveira, E.F.; Santos-Martins, D.; Bras, N.F.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Receptor-based virtual screening protocol for drug discovery. Arch. Biochem. Biophys. 2015, 582, 56–67. [Google Scholar] [PubMed]
- Channar, P.A.; Saeed, A.; Shahzad, D.; Larik, F.A.; Hassan, M.; Raza, H.; Abbas, Q.; Seo, S.Y. Extending the scope of amantadine drug by incorporation of phenolic azoschiff bases as potent selective inhibitors of carbonic anhydrase II, drug-likeness and binding analysis. Chem. Biol. Drug Des. 2018, 92, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Qazi, S.U.; Rahman, S.U.; Awan, A.N.; Al-Rashida, M.; Alharthy, R.D.; Asari, A.; Hameed, A.; Iqbal, J. Semicarbazone derivatives as urease inhibitors: Synthesis, biological evaluation, molecular docking studies and in-silico ADME evaluation. Bioorg. Chem. 2018, 79, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Su, M.; Han, L.; Liu, J.; Yang, Q.; Li, Y.; Wang, R. Forging the basis for developing protein-ligand interaction scoring functions. Acc. Chem. Res. 2017, 50, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Su, M.; Liu, Z.; Li, J.; Li, Y.; Wang, R. Enhance the performance of current scoring functions with the aid of 3D protein-ligand interaction fingerprints. BMC Bioinform. 2017, 18, 343. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, M.; Liu, Z.; Li, J.; Liu, J.; Han, L.; Wang, R. Assessing protein-ligand interaction scoring functions with the CASF-2013 benchmark. Nat. Protoc. 2018, 13, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Y.; Han, L.; Li, J.; Liu, J.; Zhao, Z.; Nie, W.; Liu, Y.; Wang, R. PDB-wide collection of binding data: Current status of the PDBbind database. Bioinformatics 2015, 31, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Kokkonen, P.; Kokkola, T.; Suuronen, T.; Poso, A.; Jarho, E.; Lahtela-Kakkonen, M. Virtual screening approach of sirtuin inhibitors results in two new scaffolds. Eur. J. Pharm. Sci. 2015, 76, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, D.; Fakhraei, S.; Getoor, L. A probabilistic approach for collective similarity-based drug-drug interaction prediction. Bioinformatics 2016, 32, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Gao, J.; Wu, D.; Zhang, S.; Mamitsuka, H.; Zhu, S. Druge-rank: Improving drug-target interaction prediction of new candidate drugs or targets by ensemble learning to rank. Bioinformatics 2016, 32, i18–i27. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Hu, G.; Wang, K.; Brylinski, M.; Xie, L.; Kurgan, L. PDID: Database of molecular-level putative protein-drug interactions in the structural human proteome. Bioinformatics 2016, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Li, W.; Sun, H.Z.; Zhou, Y.; Huang, N. computational chemistry in structure-based drug design. Yao Xue Xue Bao 2013, 48, 1041–1052. [Google Scholar] [PubMed]
- Bower, R.L.; Hay, D.L. Amylin structure-function relationships and receptor pharmacology: Implications for amylin mimetic drug development. Br. J. Pharmacol. 2016, 173, 1883–1898. [Google Scholar] [CrossRef] [PubMed]
- Drinkwater, N.; McGowan, S. From crystal to compound: Structure-based antimalarial drug discovery. Biochem. J. 2014, 461, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Bemis, G.W.; Murcko, M.A. Properties of known drugs. 2. Side chains. J. Med. Chem. 1999, 42, 5095–5099. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.K.; Verma, S.; Prabhakar, Y.S. Topological and physicochemical characteristics of 1,2,3,4-tetrahydroacridin-9(10h)-ones and their antimalarial profiles: A composite insight to the structure-activity relation. Curr. Comput. Aided Drug Des. 2013, 9, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Fattah, T.A.; Saeed, A.; Channar, P.A.; Larik, F.A.; Hassan, M.; Raza, H.; Abbas, Q.; Seo, S.Y. Synthesis and molecular docking studies of (e)-4-(substituted-benzylideneamino)-2h-chromen-2-one derivatives: Entry to new carbonic anhydrase class of inhibitors. Drug Res. 2018, 68, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hou, T. Drug and drug candidate building block analysis. J. Chem. Inf. Model. 2010, 50, 55–67. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Jia, C.; Duan, Y.; Zou, Q. 70propred: A predictor for discovering sigma70 promoters based on combining multiple features. BMC Syst. Biol. 2018, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Mignani, S.; Rodrigues, J.; Tomas, H.; Jalal, R.; Singh, P.P.; Majoral, J.P.; Vishwakarma, R.A. Present drug-likeness filters in medicinal chemistry during the hit and lead optimization process: How far can they be simplified? Drug Discov. Today 2018, 23, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Starosyla, S.A.; Volynets, G.P.; Bdzhola, V.G.; Golub, A.G.; Protopopov, M.V.; Yarmoluk, S.M. Ask1 pharmacophore model derived from diverse classes of inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 4418–4423. [Google Scholar] [CrossRef] [PubMed]
- Vrontaki, E.; Melagraki, G.; Voskou, S.; Phylactides, M.S.; Mavromoustakos, T.; Kleanthous, M.; Afantitis, A. Development of a predictive pharmacophore model and a 3D-QSAR study for an in silico screening of new potent BCR-ABL kinase inhibitors. Mini Rev. Med. Chem. 2017, 17, 188–204. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Hu, B.; Wang, J.; Zhu, F.; Kang, Y.; Li, D.; Sun, H.; Kong, D.X.; Hou, T. Cheminformatic insight into the differences between terrestrial and marine originated natural products. J. Chem. Inf. Model. 2018, 58, 1182–1193. [Google Scholar] [CrossRef] [PubMed]
- Granter, S.R.; Beck, A.H.; Papke, D.J., Jr. Alphago, deep learning, and the future of the human microscopist. Arch. Pathol. Lab. Med. 2017, 141, 619–621. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Park, J.; Bong, K.; Shin, D.; Lee, J.; Choi, S.; Yoo, H.J. An energy-efficient and scalable deep learning/inference processor with tetra-parallel MIMD architecture for big data applications. IEEE Trans. Biomed. Circuits Syst. 2015, 9, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Peng, M.M.; Liao, B.; Huang, G.H.; Li, W.B.; Xie, D.F. The advances and challenges of deep learning application in biological big data processing. Curr. Bioinform. 2018, 13, 352–359. [Google Scholar] [CrossRef]
- Wei, L.; Ding, Y.; Su, R.; Tang, J.; Zou, Q. Prediction of human protein subcellular localization using deep learning. J. Parallel Distr. Comput. 2018, 117, 212–217. [Google Scholar] [CrossRef]
- Ranjan, P.; Athar, M.; Jha, P.C.; Krishna, K.V. Probing the opportunities for designing anthelmintic leads by sub-structural topology-based QSAR modelling. Mol. Diver. 2018, 22, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Li, J.; Hong, Q.; Lin, Z.; Wu, Y.; Shi, H.; Ju, Y. Prediction of microRNA-disease associations based on social network analysis methods. BioMed Res. Int. 2015, 2015, 810514. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Zuo, Y.; Zou, Q. O-glcnacpred-II: An integrated classification algorithm for identifying o-glcnacylation sites based on fuzzy undersampling and a k-means PCA oversampling technique. Bioinformatics 2018, 34, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Jia, C.; Zou, Q. 4mCPred: Machine learning methods for DNA n4-methylcytosine sites prediction. Bioinformatics 2018. [Google Scholar] [CrossRef] [PubMed]
- MotieGhader, H.; Gharaghani, S.; Masoudi-Sobhanzadeh, Y.; Masoudi-Nejad, A. Sequential and mixed genetic algorithm and learning automata (SGALA, MGALA) for feature selection in QSAR. Iran. J. Pharm. Res. 2017, 16, 533–553. [Google Scholar] [PubMed]
- Ekins, S.; de Siqueira-Neto, J.L.; McCall, L.I.; Sarker, M.; Yadav, M.; Ponder, E.L.; Kallel, E.A.; Kellar, D.; Chen, S.; Arkin, M.; et al. Machine learning models and pathway genome data base for trypanosomacruzi drug discovery. PLoS Negl. Trop. Dis. 2015, 9, e0003878. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.; Balbas-Martinez, V.; Jergens, A.E.; Troconiz, I.F.; Allenspach, K.; Mochel, J.P. Model-based reverse translation between veterinary and human medicine: The one health initiative. CPT Pharmacometr. Syst. Pharmacol. 2018, 7, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Yosipof, A.; Guedes, R.C.; Garcia-Sosa, A.T. Data mining and machine learning models for predicting drug likeness and their disease or organ category. Front. Chem. 2018, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Ning, Z.; Hu, D.; Zhang, M.; Zhao, L.; Lin, C.; Zhong, L.L.D.; Yang, Z.; Xu, H.; Bian, Z. Uncovering the mechanisms of Chinese herbal medicine (MaZiRenWan) for functional constipation by focused network pharmacology approach. Front. Pharmacol. 2018, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, J.; Wu, Z.; Huang, C.; Lu, A.; Wang, Y. Systems pharmacology exploration of botanic drug pairs reveals the mechanism for treating different diseases. Sci. Rep. 2016, 6, 36985. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Subbiah, K. Maximizing lipocalin prediction through balanced and diversified training set and decision fusion. Comput. Biol. Chem. 2015, 59, 101–110. [Google Scholar] [CrossRef] [PubMed]


| Author | Title | Application | Ref. |
|---|---|---|---|
| Li, X.; Kang, H.; Liu, W.; Singhal, S.; Jiao, N.; Wang, Y.; Zhu, L.; Zhu, R. | In silico design of novel proton-pump inhibitors with reduced adverse effects. | Virtual screening is used to select molecules with the desired pKa values | [19] |
| Azad, I.; Nasibullah, M.; Khan, T.; Hassan, F.; Akhter, Y. | Exploring the novel heterocyclic derivatives as lead molecules for design and development of potent anticancer agents. | Used a novel heterocyclic derivative as a lead compound to perform virtual screening on 27 compounds previously screened to develop an effective anticancer drug | [20] |
| McKerrow, J.H.; Lipinski, C.A. | The rule of five should not impede anti-parasitic drug development. | Proposed five rules for drug-like | [27] |
| Fong, P.; Ao, C.N.; Tou, K.I.; Huang, K.M.; Cheong, C.C.; Meng, L.R | Experimental and in silico analysis of cordycepin and its derivatives as endometrial cancer treatment. | Used molecular docking scores to evaluate the possibility of cordycepin and its derivatives as therapeutic agents for endometrial cancer | [28] |
| Ai, H.; Wu, X.; Qi, M.; Zhang, L.; Hu, H.; Zhao, Q.; Zhao, J.; Liu, H. | Study on the mechanisms of active compounds in traditional Chinese medicine for the treatment of influenza virus by virtual screening. | The study utilizes structure-based molecular docking techniques to screen for more than 10,000 molecular structures | [29] |
| Onawole, A.T.; Kolapo, T.U.; Sulaiman, K.O.; Adegoke, R.O. | Structure based virtual screening of the ebola virus trimeric glycoprotein using consensus scoring. | Used the co-product score method to evaluate the three drug candidates currently selected for the treatment of the Ebola virus | [30] |
| Author | Title | Application | Ref. |
|---|---|---|---|
| Bemis, G.W.; Murcko, M.A. | Properties of known drugs. 2. Side chains. | found a large amount of information available in the corresponding analysis of the molecular structure of drugs | [63] |
| Wang, J.; Hou, T. | Drug and drug candidate building block analysis. | combined statistically relevant methods to compare a variety of predicted molecules with known drug molecules for more comprehensive attributes | [66] |
| Starosyla, S.A.; Volynets, G.P.; Bdzhola, V.G.; Golub, A.G.; Protopopov, M.V.; Yarmoluk, S.M. | Ask1 pharmacophore model derived from diverse classes of inhibitors. | The location of the pharmacophore features in the model corresponds to the conformation of the ASK1 high activity inhibitor, which interacts with the binding site of ASK1 | [69] |
| Shang, J.; Hu, B.; Wang, J.; Zhu, F.; Kang, Y.; Li, D.; Sun, H.; Kong, D.X.; Hou, T. | Cheminformatic insight into the differences between terrestrial and marine originated natural products. | used chemical informatics to study the physical and chemical structures and pharmacological pharmacophores of terrestrial and marine natural products | [71] |
| Author | Title | Application | Ref. |
|---|---|---|---|
| Ekins, S.; de Siqueira-Neto, J.L.; McCall, L.I.; Sarker, M.; Yadav, M.; Ponder, E.L.; Kallel, E.A.; Kellar, D.; Chen, S.; Arkin, M. | Machine learning models and pathway genome data base for trypanosomacruzi drug discovery. | developed a Bayesian machine learning model for screening active compounds for the treatment of neural tube defects caused by trypanosomacruzi | [81] |
| Schneider, B.; Balbas-Martinez, V.; Jergens, A.E.; Troconiz, I.F.; Allenspach, K.; Mochel, J.P. | Model-based reverse translation between veterinary and human medicine: The one health initiative. | have established a network model based on recursive partitioning algorithms based on 3117 drugs and 2238 non-pharmaceuticals, but the effect is not particularly ideal | [82] |
| Yosipof, A.; Guedes, R.C.; Garcia-Sosa, A.T. | Data mining and machine learning models for predicting drug likeness and their disease or organ category. | used a variety of different machine learning methods to form a new integrated learning method called AL Boost | [83] |
| Huang, T.; Ning, Z.; Hu, D.; Zhang, M.; Zhao, L.; Lin, C.; Zhong, L.L.D.; Yang, Z.; Xu, H.; Bian, Z. | Uncovering the mechanisms of Chinese herbal medicine (mazirenwan) for functional constipation by focused network pharmacology approach. | The method incorporates a variety of machine learning methods for predicting possible targets for representative compounds | [84] |
| Zhou, W.; Wang, J.; Wu, Z.; Huang, C.; Lu, A.; Wang, Y. | Systems pharmacology exploration of botanic drug pairs reveals the mechanism for treating different diseases. | used a machine-based C-P network analysis and C-P-T network analysis method to predict and analyze the extracted active compounds | [85] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.; Zhang, L.; Wang, M.; Zhang, R.; Wang, C.; Zhang, C. Prediction Methods of Herbal Compounds in Chinese Medicinal Herbs. Molecules 2018, 23, 2303. https://doi.org/10.3390/molecules23092303
Han K, Zhang L, Wang M, Zhang R, Wang C, Zhang C. Prediction Methods of Herbal Compounds in Chinese Medicinal Herbs. Molecules. 2018; 23(9):2303. https://doi.org/10.3390/molecules23092303
Chicago/Turabian StyleHan, Ke, Lei Zhang, Miao Wang, Rui Zhang, Chunyu Wang, and Chengzhi Zhang. 2018. "Prediction Methods of Herbal Compounds in Chinese Medicinal Herbs" Molecules 23, no. 9: 2303. https://doi.org/10.3390/molecules23092303
APA StyleHan, K., Zhang, L., Wang, M., Zhang, R., Wang, C., & Zhang, C. (2018). Prediction Methods of Herbal Compounds in Chinese Medicinal Herbs. Molecules, 23(9), 2303. https://doi.org/10.3390/molecules23092303







