Synthesis and Antisense Properties of 2′β-F-Arabinouridine Modified Oligonucleotides with 4′-C-OMe Substituent
Abstract
:1. Introduction
2. Results and Discussion
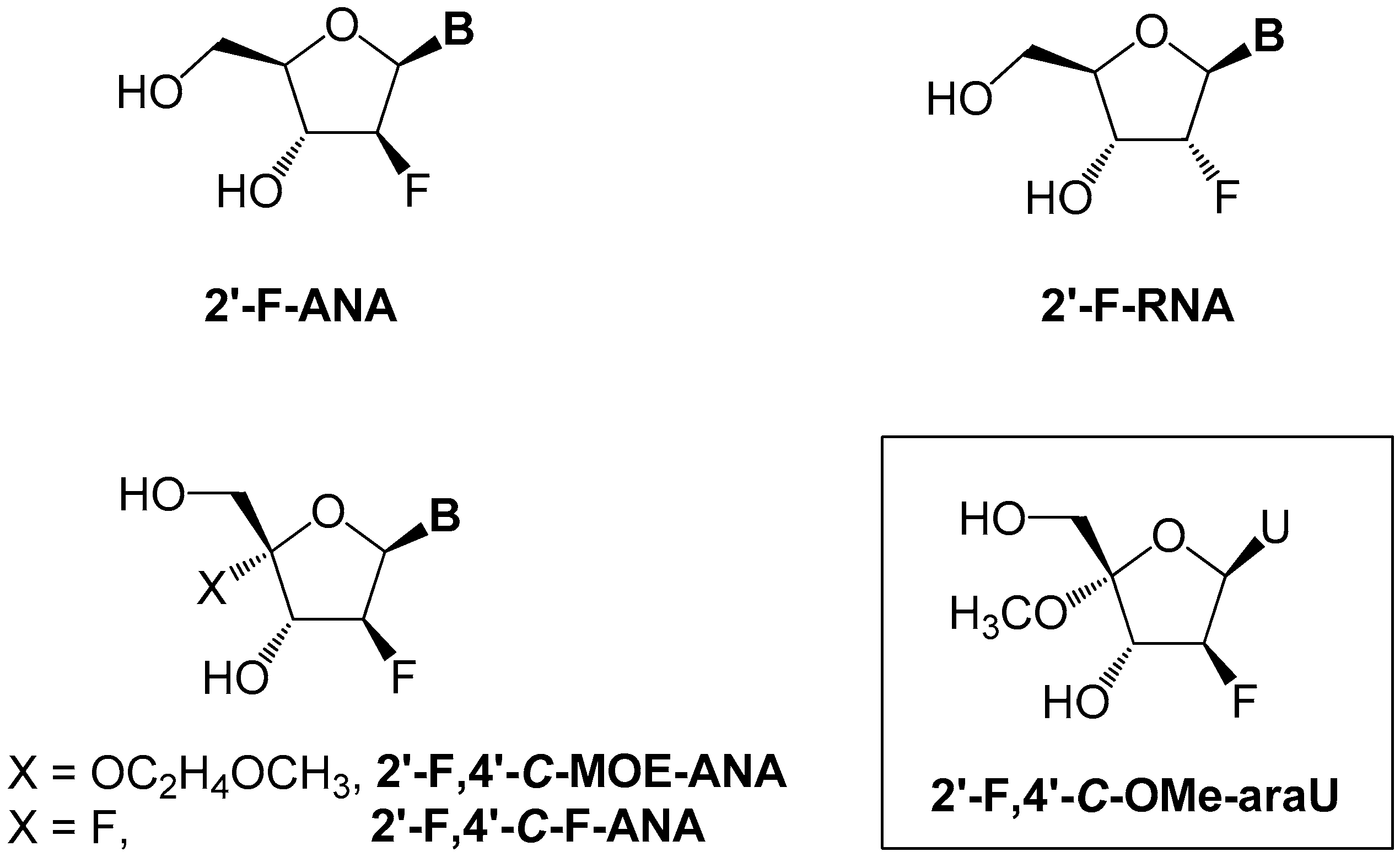
2.1. Chemistry
2.2. Hybridization Properties
2.3. Nuclease Stability
3. Conclusions
4. General Experimental Procedure
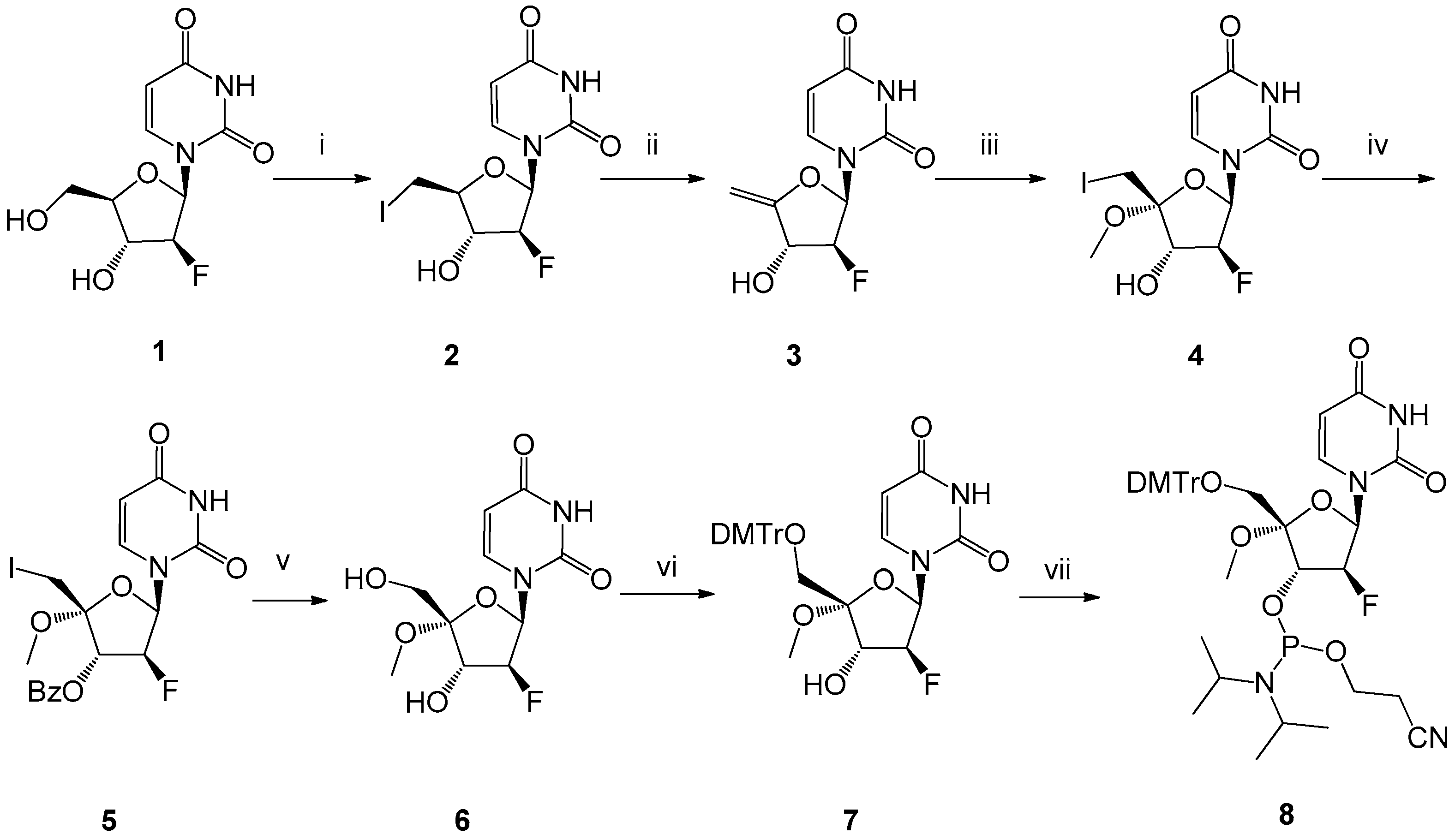
4.1. Chemistry
4.1.1. 2′,5′-Dideoxy-5′-iodo-2′-fluoro-β-d-arabinouridine (2)
4.1.2. 2′,5′-Dideoxy-2′-fluoro-4′-methylene-β-d-arabinouridine (3)
4.1.3. 2′,5′-Dideoxy-2′-fluoro-5′-iodo-4′-C-methoxy-β-d-arabinouridine (4)
4.1.4. 3′-O-Benzoyl-2′,5′-dideoxy-2′-fluoro-5′-iodo-4′-C-methoxy-β-d-arabinouridine (5)
4.1.5. 2′-Deoxy-2′-fluoro-4′-C-methoxy-β-d-arabinouridine (6)
4.1.6. 2′-Deoxy-2′-fluoro-5′-O-(4,4′-dimethoxy)trityl-4′-C-methoxy-β-d-arabinouridine (7)
4.1.7. 2′-Deoxy-5′-O-(4,4′-dimethoxy)trityl-4′-C-methoxy-β-d-arabinouridine-3′-O-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (8)
4.1.8. Oligonucleotide Synthesis
4.2. Ultraviolet (UV) Melting Experiments
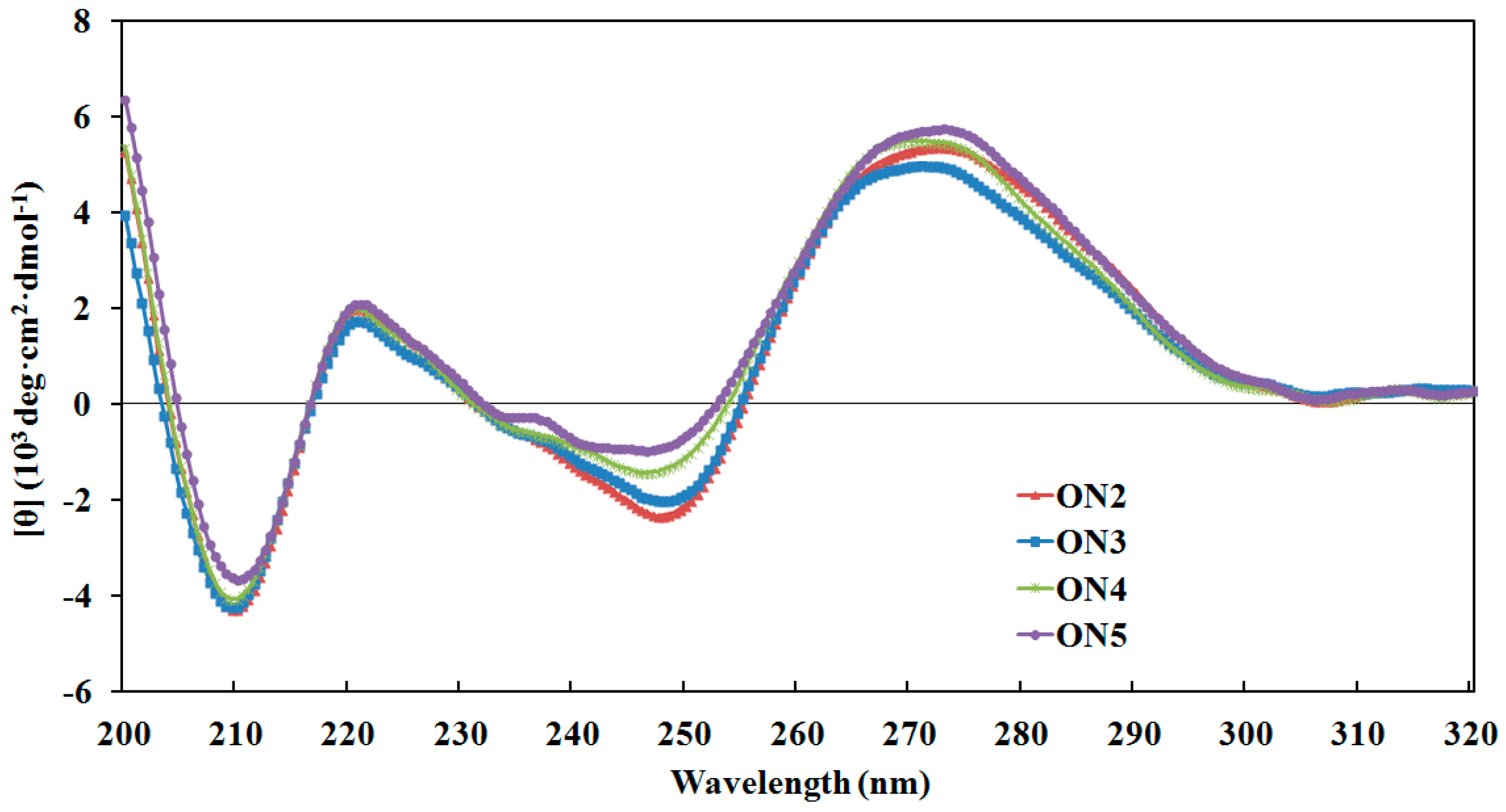
4.3. Circular Dichroism Spectroscopy
4.4. Nuclease Stability Experiments
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [PubMed]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [PubMed]
- Dirin, M.; Winkler, J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2013, 13, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, M.E.; Dwight, T.; Berdeja, A.; Swayze, E.E.; Jung, M.E.; Seth, P.P. Comparison of duplex stabilizing properties of 2′-fluorinated nucleic acid analogues with furanose and non-Furanose sugar rings. J. Org. Chem. 2014, 79, 8877–8881. [Google Scholar] [CrossRef] [PubMed]
- Ellipilli, S.; Ganesh, K.N. Fluorous peptide nucleic acids: PNA analogues with fluorine in backbone (γ-CF2-apg-PNA) enhance cellular uptake. J. Org. Chem. 2015, 80, 9185–9191. [Google Scholar] [PubMed]
- Dugovic, B.; Leumann, C.J. A 6′-fluoro-substituent in bicyclo-DNA increases affinity to complementary RNA presumably by CF–HC pseudohydrogen bonds. J. Org. Chem. 2014, 79, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Hagmann, W.K. The many roles for fluorine in medicinal chemistry. J. Med. Chem. 2008, 51, 4359–4369. [Google Scholar] [CrossRef] [PubMed]
- Martínpintado, N.; Yahyaeeanzahaee, M.; Deleavey, G.F.; Portella, G.; Orozco, M.; Damha, M.J.; González, C. Dramatic effect of furanose C2′ substitution on structure and stability: Directing the folding of the human telomeric quadruplex with a single fluorine atom. J. Am. Chem. Soc. 2013, 135, 5344–5347. [Google Scholar]
- Anzahaee, M.Y.; Watts, J.K.; Alla, N.R.; Nicholson, A.W.; Damha, M.J. Energetically important C-H⋯F-C pseudohydrogen bonding in water: Evidence and application to rational design of oligonucleotides with high binding affinity. J. Am. Chem. Soc. 2011, 133, 728–731. [Google Scholar] [PubMed]
- Gore, K.R.; Harikrishna, S.; Pradeepkumar, P. Influence of 2′-Fluoro versus 2′-O-Methyl Substituent on the Sugar Puckering of 4′-C-Aminomethyluridine. J. Org. Chem. 2013, 78, 9956–9962. [Google Scholar] [PubMed]
- Watts, J.K.; Choubdar, N.; Sadalapure, K.; Robert, F.; Wahba, A.S.; Pelletier, J.; Pinto, B.M.; Damha, M.J. 2′-Fluoro-4′-thioarabino-modified oligonucleotides: Conformational switches linked to siRNA activity. Nucleic Acids Res. 2007, 35, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.K.; Damha, M.J. 2′F-Arabinonucleic acids (2′F-ANA)-history, properties, and new frontiers. Can. J. Chem. 2008, 86, 641–656. [Google Scholar] [CrossRef]
- Watts, J.K.; Martínpintado, N.; Gómezpinto, I.; Schwartzentruber, J.; Portella, G.; Orozco, M.; González, C.; Damha, M.J. Differential stability of 2′F-ANA•RNA and ANA•RNA hybrid duplexes: Roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010, 38, 2498–2511. [Google Scholar] [CrossRef] [PubMed]
- Martínpintado, N.; Yahyaeeanzahaee, M.; Camposolivas, R.; Noronha, A.M.; Wilds, C.J.; Damha, M.J.; González, C. The solution structure of double helical arabino nucleic acids (ANA and 2′F-ANA): Effect of arabinoses in duplex-hairpin interconversion. Nucleic Acids Res. 2012, 40, 9329–9339. [Google Scholar] [CrossRef] [PubMed]
- Liboska, R.; Snášel, J.; Barvík, I.; Buděšínský, M.; Pohl, R.; Točík, Z.; Páv, O.; Rejman, D.; Novák, P.; Rosenberg, I. 4′-Alkoxy oligodeoxynucleotides: A novel class of RNA mimics. Org. Biomol. Chem. 2011, 9, 8261–8267. [Google Scholar] [CrossRef] [PubMed]
- Petrová, M.; Páv, O.; Buděšínský, M.; Zborníková, E.; Novák, P.; Rosenbergová, Š.; Pačes, O.; Liboska, R.; Dvořáková, I.; Simak, O.; et al. Straightforward synthesis of purine 4′-alkoxy-2′-deoxynucleosides: First report of mixed purine-pyrimidine 4′-alkoxyoligodeoxynucleotides as new RNA mimics. Org. Lett. 2015, 17, 3426–3429. [Google Scholar] [CrossRef] [PubMed]
- Martinezmontero, S.; Deleavey, G.F.; Dierkerviik, A.; Lindovska, P.; Ilina, T.; Portella, G.; Orozco, M.; Parniak, M.A.; González, C.; Damha, M.J. Synthesis and properties of 2′-Deoxy-2′,4′-difluoroarabinose modified nucleic acids (2′,4′-diFANA). J. Org. Chem. 2015, 80, 3083–3091. [Google Scholar] [CrossRef] [PubMed]
- Gore, K.R.; Nawale, G.N.; Harikrishna, S.; Chittoor, V.G.; Pandey, S.K.; Höbartner, C.; Patankar, S.; Pradeepkumar, P.I. Synthesis, gene silencing, and molecular modeling studies of 4′-C-aminomethyl-2′-O-methyl modified small interfering RNAs. J. Org. Chem. 2012, 77, 3233–3245. [Google Scholar] [CrossRef] [PubMed]
- Malek-Adamian, E.; Guenther, D.C.; Matsuda, S.; Martínez-Montero, S.; Zlatev, I.; Harp, J.; Burai Patrascu, M.; Foster, D.J.; Fakhoury, J.; Perkins, L.; et al. 4′-C-Methoxy-2′-deoxy-2′-fluoro modified ribonucleotides improve metabolic stability and elicit efficient RNAi-mediated gene silencing. J. Am. Chem. Soc. 2017, 139, 14542–14555. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wang, J.; Lu, D.-D.; Yang, J.; Qin, B.-J.; He, X.-Y.; Wang, S.-Q. Synthesis and antisense properties of 2′-β-F- and 2′-α-F-2′-deoxy-uridines modified oligonucleotides with 4′-C-(2-methoxyethoxy) substituent. Acta Pharm. Sin. 2016, 51, 1271–1280. [Google Scholar]
- Wang, G.; Deval, J.; Hong, J.; Dyatkina, N.; Prhavc, M.; Taylor, J.; Fung, A.; Jin, Z.; Stevens, S.K.; Serebryany, V. Discovery of 4′-chloromethyl-2′-deoxy-3′,5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 2015, 58, 1862–1878. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

| # | Sequence (5′–3′) | ssRNA | ssDNA | RNA Selectivity | ||
|---|---|---|---|---|---|---|
| Tm | ΔTm | Tm | ΔTm b | ΔTm (RNA) − ΔTm (DNA) | ||
| ON1 | GCGTTTTTTGCT | 47.5 | 51.7 | |||
| ON2 | GCGTTXTTTGCT | 47.0 | −0.5 | 48.4 | −3.3 | 2.8 |
| ON3 | GCGTTYTTTGCT | 46.3 | −1.2 | 51.2 | −0.5 | −0.7 |
| ON4 | GCGTTXTXTGCT | 46.3 | −1.2 | 46.1 | −5.6 | 4.4 |
| ON5 | GCGXTXTXTGCT | 42.9 | −4.6 | ND c | ||
| ON6 | GCGTTGTTTGCT | 50.4 | 56.6 | |||
| ON7 | GCGTXGTTTGCT | 52.0 | 1.6 | 52.5 | −4.1 | 5.7 |
| ON8 | GCGTTGXTTGCT | 49.5 | −0.9 | 51.2 | −5.4 | 4.5 |
| ON9 | GCGTTATTTGCT | 45.6 | 51.3 | |||
| ON10 | GCGTXATTTGCT | 48.2 | 2.6 | 47.9 | −3.4 | 6.0 |
| ON11 | GCGTTAXTTGCT | 44.6 | −1.0 | 46.5 | −4.8 | 3.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.-Y.; Wang, J.; Lu, D.-D.; Wang, S.-Q. Synthesis and Antisense Properties of 2′β-F-Arabinouridine Modified Oligonucleotides with 4′-C-OMe Substituent. Molecules 2018, 23, 2374. https://doi.org/10.3390/molecules23092374
He X-Y, Wang J, Lu D-D, Wang S-Q. Synthesis and Antisense Properties of 2′β-F-Arabinouridine Modified Oligonucleotides with 4′-C-OMe Substituent. Molecules. 2018; 23(9):2374. https://doi.org/10.3390/molecules23092374
Chicago/Turabian StyleHe, Xiao-Yang, Jing Wang, Dan-Dan Lu, and Sheng-Qi Wang. 2018. "Synthesis and Antisense Properties of 2′β-F-Arabinouridine Modified Oligonucleotides with 4′-C-OMe Substituent" Molecules 23, no. 9: 2374. https://doi.org/10.3390/molecules23092374
APA StyleHe, X.-Y., Wang, J., Lu, D.-D., & Wang, S.-Q. (2018). Synthesis and Antisense Properties of 2′β-F-Arabinouridine Modified Oligonucleotides with 4′-C-OMe Substituent. Molecules, 23(9), 2374. https://doi.org/10.3390/molecules23092374






