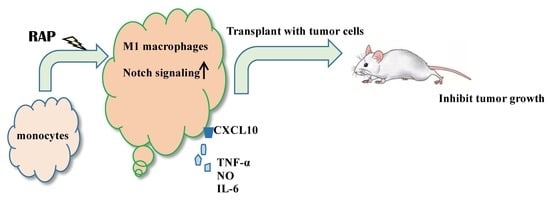
Astragalus Polysaccharide RAP Induces Macrophage Phenotype Polarization to M1 via the Notch Signaling Pathway
Abstract
:1. Introduction
2. Results
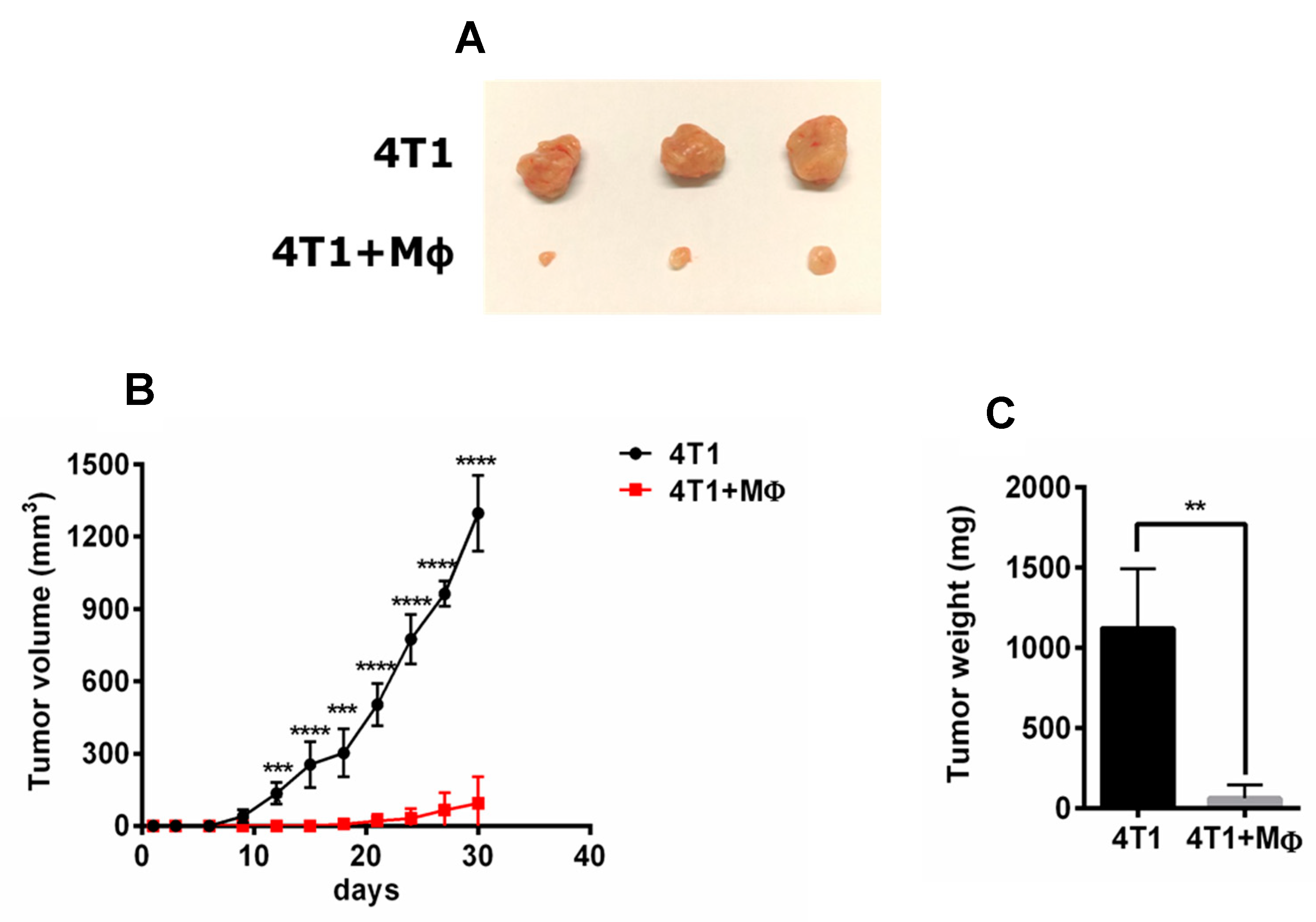
2.1. RAP-Stimulated BMDMs Decreased Tumor Volume and Tumor Weight
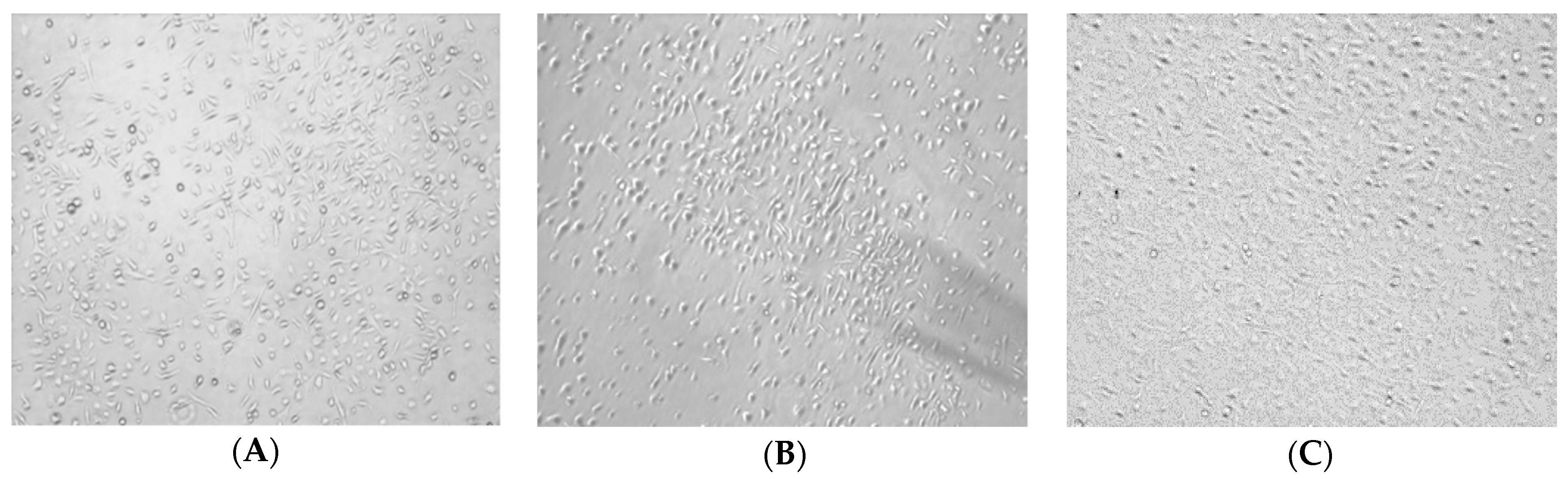
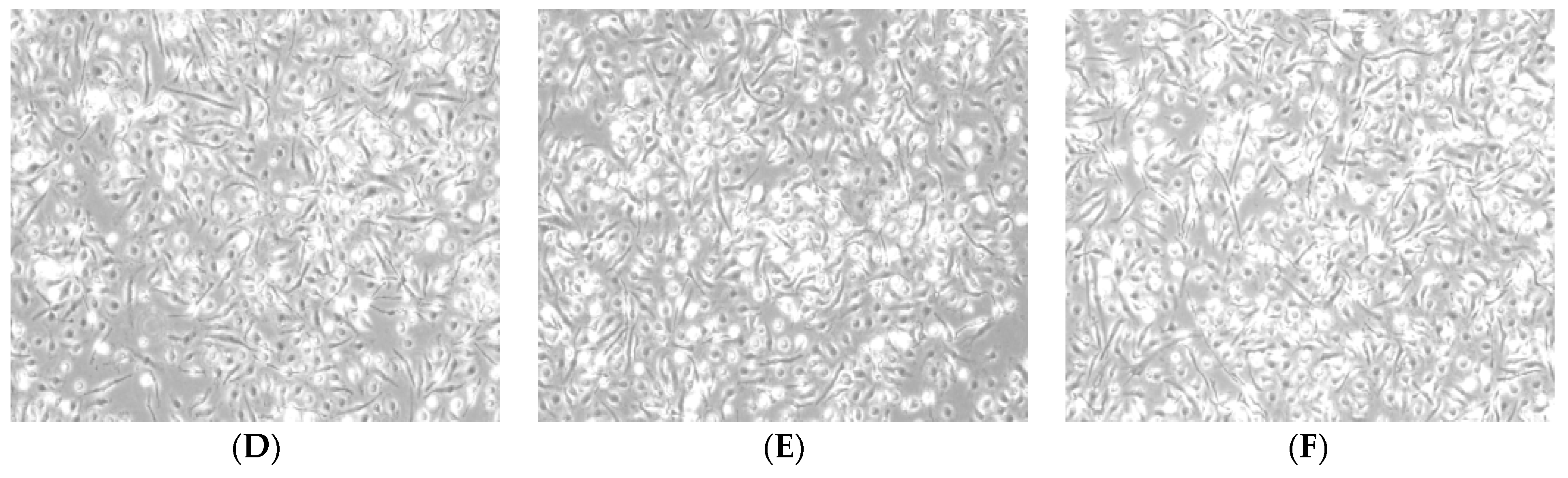
2.2. Morphology of BMDMs Induced by RAP
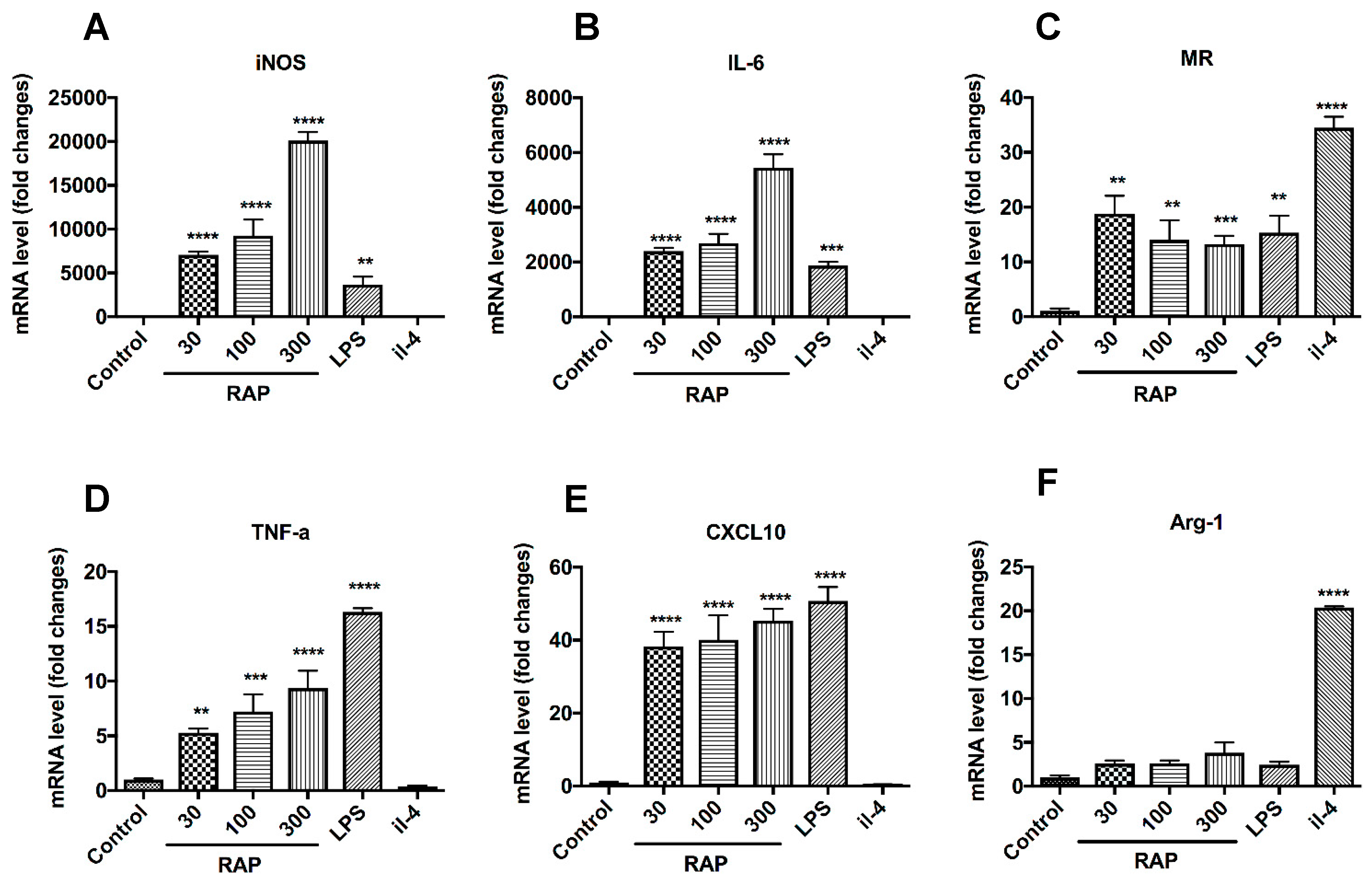
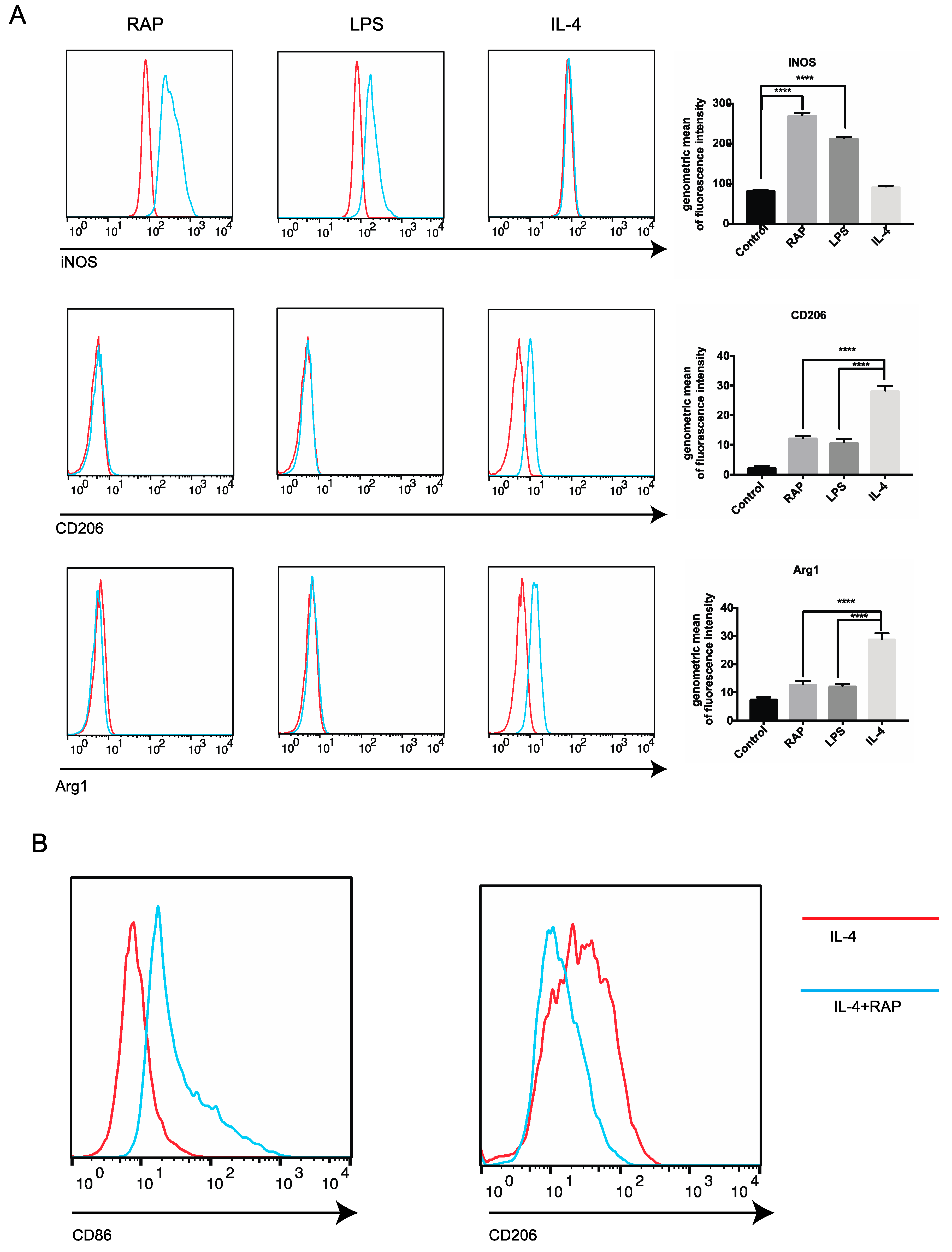
2.3. Analysis of M1 Marker Expression on BMDMs’ Surface
2.4. Gene Expression of the Notch Signaling Pathway Induced by RAP
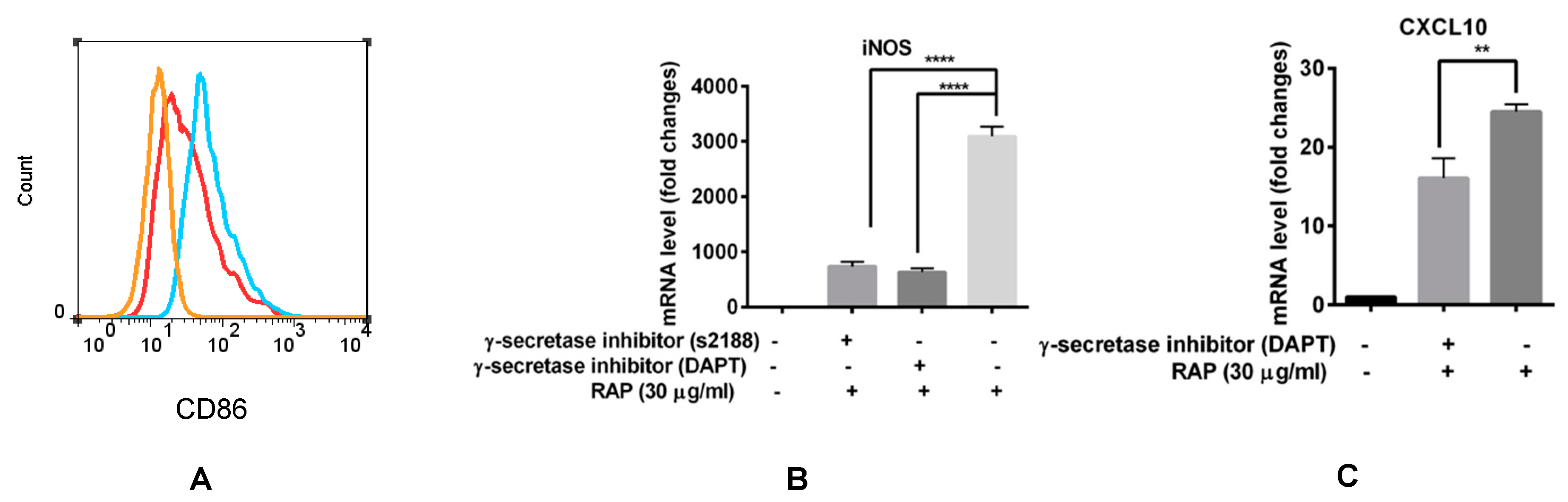
2.5. Blocking of the Notch Signaling Pathway Results in M1 Marker Decrease even in the Presence of RAP
3. Materials and Methods
3.1. Materials
3.1.1. Materials
3.1.2. Cells Cultures and Animals
3.1.3. Mouse Models
3.2. Analysis of Macrophage Surface Antigen Expression by Flow Cytometry
3.3. Cell Morphology
3.4. RNA Isolation and qRT-PCR
3.5. Western Blot Analysis
3.6. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jin, M.; Zhao, K.; Huang, Q.; Shang, P. Structural features and biological activities of the polysaccharides from Astragalus membranaceus. Int. J. Biol. Macromol. 2014, 64, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.-H.; Ma, Z.-X.; Zhu, J.; Yu, X.-H.; Weng, D.-P. Characterization of polysaccharide from Astragalus radix as the macrophage stimulator. Cell. Immunol. 2011, 271, 329–334. [Google Scholar] [CrossRef]
- Li, R.; Chen, W.-C.; Wang, W.-P.; Tian, W.-Y.; Zhang, X.-G. Antioxidant activity of Astragalus polysaccharides and antitumour activity of the polysaccharides and siRNA. Carbohydr. Polym. 2010, 82, 240–244. [Google Scholar] [CrossRef]
- Li, X.-T.; Zhang, Y.-K.; Kuang, H.-X.; Jin, F.-X.; Liu, D.-W.; Gao, M.-B.; Liu, Z.; Xin, X.-J. Mitochondrial protection and anti-aging activity of Astragalus polysaccharides and their potential mechanism. Int. J. Mol. Sci. 2012, 13, 1747–1761. [Google Scholar] [CrossRef]
- Niu, Y.; Wang, H.; Xie, Z.; Whent, M.; Gao, X.; Zhang, X.; Zou, S.; Yao, W.; Yu, L. Structural analysis and bioactivity of a polysaccharide from the roots of Astragalus membranaceus (Fisch) Bge. var. mongolicus (Bge.) Hsiao. Food Chem. 2011, 128, 620–626. [Google Scholar] [CrossRef]
- Liu, M.; Wu, K.; Mao, X.; Wu, Y.; Ouyang, J. Astragalus polysaccharide improves insulin sensitivity in KKAy mice: Regulation of PKB/GLUT4 signaling in skeletal muscle. J. Ethnopharmacol. 2010, 127, 32–37. [Google Scholar] [CrossRef]
- Mao, X.-Q.; Yu, F.; Wang, N.; Wu, Y.; Zou, F.; Wu, K.; Liu, M.; Ouyang, J.-P. Hypoglycemic effect of polysaccharide enriched extract of Astragalus membranaceus in diet induced insulin resistant C57BL/6J mice and its potential mechanism. Phytomedicine 2009, 16, 416–425. [Google Scholar] [CrossRef]
- He, X.; Shu, J.; Xu, L.; Lu, C.; Lu, A. Inhibitory effect of Astragalus polysaccharides on lipopolysaccharide-induced TNF-a and IL-1β production in THP-1 cells. Molecules 2012, 17, 3155–3164. [Google Scholar] [CrossRef]
- Lu, J.; Chen, X.; Zhang, Y.; Xu, J.; Zhang, L.; Li, Z.; Liu, W.; Ouyang, J.; Han, S.; He, X. Astragalus polysaccharide induces anti-inflammatory effects dependent on AMPK activity in palmitate-treated RAW264. 7 cells. Int. J. Mol. Med. 2013, 31, 1463–1470. [Google Scholar] [CrossRef]
- Huang, H.-F.; Qian, J.-Y.; Xie, S.-R. Effects of Astragalus Polysaccharide on Apoptosis and Cell Cycle of Human Gastric Carcinoma Cell Line MKN45. J. Clin. Med. Pract. 2010, 19, 006. [Google Scholar]
- Li, L.-K.; Huang, Y.-F.; Xie, H.-H.; Chen, G.; Wang, B.-R.; Kuang, W.-J.; Wan, L.-H. Effects of Astragalus Injection on the Protein Expression of Bax and Bcl-2 in H22 Tumor Cells Bearing Mice. Chin. J. Exp. Tradit. Med. Formulae 2011, 17, 055. [Google Scholar]
- Li, J.; Bao, Y.; Zhu, X.; Liu, J.; Wang, H. Immunoregulatory activity of polysaccharopeptide and Astragalus polysaccharides on EAC tumor-bearing mice. Chin. J. Chin. Mater. Med. 2008, 33, 924–927. [Google Scholar]
- Li, R.; Chen, W.-C.; Wang, W.-P.; Tian, W.-Y.; Zhang, X.-G. Extraction, characterization of Astragalus polysaccharides and its immune modulating activities in rats with gastric cancer. Carbohydr. Polym. 2009, 78, 738–742. [Google Scholar] [CrossRef]
- Tian, Q.-E.; Li, H.-D.; Miao, Y.; Cai, H.-L.; Tan, Q.-Y.; Zhang, W.-Y. Astragalus polysaccharides can regulate cytokine and P-glycoprotein expression in H22 tumor-bearing mice. Word J. Gastroenterol. 2012, 18, 079. [Google Scholar] [CrossRef]
- Yin, J.-Y.; Chan, B.C.-L.; Yu, H.; Lau, I.Y.-K.; Han, X.-Q.; Cheng, S.-W.; Wong, C.-K.; Bik-San Lau, C.; Xie, M.-Y.; Fung, K.-P. Separation, structure characterization, conformation and immunomodulating effect of a hyperbranched heteroglycan from Radix Astragali. Carbohydr. Polym. 2012, 87, 667–675. [Google Scholar] [CrossRef]
- Wei, W.; Xiao, H.-T.; Bao, W.-R.; Ma, D.-L.; Leung, C.-H.; Han, X.-Q.; Ko, C.-H.; Bik-San Lau, C.; Chun-Kwok, W.; Fung, K.-P. TLR-4 may mediate signaling pathways of Astragalus polysaccharide RAP induced cytokine expression of RAW264. 7 cells. J. Ethnopharmacol. 2016, 179, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, L.; Zhu, X.; Wang, Y.; Liu, W.; Gong, W. Polysaccharide Agaricus blazei Murill stimulates myeloid derived suppressor cell differentiation from M2 to M1 type, which mediates inhibition of tumour immune-evasion via the Toll-like receptor 2 pathway. Immunology 2015, 146, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Locati, M. Macrophage polarization comes of age. Immunity. 2005, 23, 344–346. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Menen, R.S.; Hassanein, M.K.; Momiyama, M.; Suetsugu, A.; Moossa, A.R.; Hoffman, R.M.; Bouvet, M. Tumor-educated macrophages promote tumor growth and peritoneal metastasis in an orthotopic nude mouse model of human pancreatic cancer. In Vivo 2012, 26, 565–569. [Google Scholar]
- Talmadge, J.E.; Donkor, M.; Scholar, E. Inflammatory cell infiltration of tumors: Jekyll or Hyde. Cancer Metastasis Rev. 2007, 26, 373–400. [Google Scholar] [CrossRef]
- Pollard, J.W. Macrophages define the invasive microenvironment in breast cancer. J. Leukoc. Biol. 2008, 84, 623–630. [Google Scholar] [CrossRef] [Green Version]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef]
- Wang, Y.-C.; He, F.; Feng, F.; Liu, X.-W.; Dong, G.-Y.; Qin, H.-Y.; Hu, X.-B.; Zheng, M.-H.; Liang, L.; Feng, L. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 2010, 70, 4840–4849. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Xiong, S. Blockade of Notch1 signaling alleviates murine lupus via blunting macrophage activation and M2b polarization. J. Immunol. 2010, 184, 6465–6478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, C.; Liu, Z.; Liu, X.; Han, C.; Cao, X.; Li, N. Notch signal suppresses Toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor κB activation. J. Biol. Chem. 2012, 287, 6208–6217. [Google Scholar] [CrossRef] [PubMed]
- Palaga, T.; Buranaruk, C.; Rengpipat, S.; Fauq, A.H.; Golde, T.E.; Kaufmann, S.H.; Osborne, B.A. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol. 2008, 38, 174–183. [Google Scholar] [CrossRef]
- Tsao, P.-N.; Wei, S.-C.; Huang, M.-T.; Lee, M.-C.; Chou, H.-C.; Chen, C.-Y.; Hsieh, W.-S. Lipopolysaccharide-induced Notch signaling activation through JNK-dependent pathway regulates inflammatory response. J. Biomed. Sci. 2011, 18, 56. [Google Scholar] [CrossRef]
- Shimizu, K.; Chiba, S.; Saito, T.; Kumano, K.; Hamada, Y.; Hirai, H. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem. Biophys. Res. Commun. 2002, 291, 775–779. [Google Scholar] [CrossRef]
- Kitamoto, T.; Takahashi, K.; Takimoto, H.; Tomizuka, K.; Hayasaka, M.; Tabira, T.; Hanaoka, K. Functional redundancy of the Notch gene family during mouse embryogenesis: Analysis of Notch gene expression in Notch3-deficient mice. Biochem. Biophys. Res. Commun. 2005, 331, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Lake, F.-R.; Noble, P.-W.; Henson, P.; Riches, D. Functional switching of macrophage responses to tumor necrosis factor-alpha (TNF alpha) by interferons. Implications for the pleiotropic activities of TNF alpha. J. Clin. Investig. 1994, 93, 1661. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Porse, B. Bone marrow-derived macrophages (BMM): Isolation and applications. Cold Spring Harb. Protoc. 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- D’Antò, V.; Eckhardt, A.; Hiller, K.-A.; Spagnuolo, G.; Valletta, R.; Ambrosio, L.; Schmalz, G.; Schweikl, H. The influence of Ni (II) on surface antigen expression in murine macrophages. Biomaterials 2009, 30, 1492–1501. [Google Scholar] [CrossRef]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2010, 10, 460. [Google Scholar] [CrossRef]
- Dalpke, A.; Heeg, K.; Bartz, H.; Baetz, A. Regulation of innate immunity by suppressor of cytokine signaling (SOCS) proteins. Immunobiology 2008, 213, 225–235. [Google Scholar] [CrossRef]
- Liu, Y.; Stewart, K.N.; Bishop, E.; Marek, C.J.; Kluth, D.C.; Rees, A.J.; Wilson, H.M. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J. Immunol. 2008, 180, 6270–6278. [Google Scholar] [CrossRef]
- Narayana, Y.; Balaji, K.-N. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J. Biol. Chem. 2008, 283, 12501–12511. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Li, Z.-P.; Bian, Z.-X.; Han, Q.-B. Astragalus Polysaccharide RAP Induces Macrophage Phenotype Polarization to M1 via the Notch Signaling Pathway. Molecules 2019, 24, 2016. https://doi.org/10.3390/molecules24102016
Wei W, Li Z-P, Bian Z-X, Han Q-B. Astragalus Polysaccharide RAP Induces Macrophage Phenotype Polarization to M1 via the Notch Signaling Pathway. Molecules. 2019; 24(10):2016. https://doi.org/10.3390/molecules24102016
Chicago/Turabian StyleWei, Wei, Zhi-Peng Li, Zhao-Xiang Bian, and Quan-Bin Han. 2019. "Astragalus Polysaccharide RAP Induces Macrophage Phenotype Polarization to M1 via the Notch Signaling Pathway" Molecules 24, no. 10: 2016. https://doi.org/10.3390/molecules24102016
APA StyleWei, W., Li, Z.-P., Bian, Z.-X., & Han, Q.-B. (2019). Astragalus Polysaccharide RAP Induces Macrophage Phenotype Polarization to M1 via the Notch Signaling Pathway. Molecules, 24(10), 2016. https://doi.org/10.3390/molecules24102016