Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network
Abstract
:1. Introduction
2. Results and Discussion
2.1. Samples
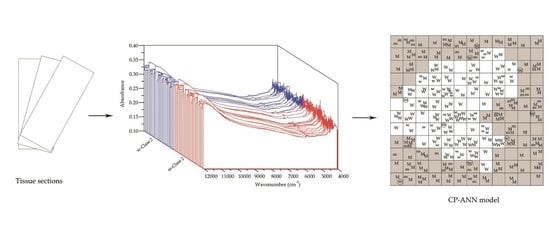
2.2. Spectral Acquisition
2.3. Data Processing
2.3.1. Selection of the Spectral Preprocessing Strategy
2.3.2. Selection of the Spectral Subrange for Modeling
2.3.3. Calibration and Validation of the CP-ANN Model
2.3.4. Diagnostic Performances of the CP-ANN Model
3. Materials and Methods
3.1. Samples
3.2. Instrument and Spectral Acquisition
3.3. Data Processing
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bouchahda, M.; Karaboué, A.; Saffroy, R.; Innominato, P.; Gorden, L.; Guettier, C.; Adam, R.; Lévi, F. Acquired KRAS mutations during progression of colorectal cancer metastases: Possible implications for therapy and prognosis. Cancer Chemother. Pharmacol. 2010, 66, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Hesari, A.R.; Khazaei, M.; Hassanian, S.M.; Ferns, G.; Avan, A. The therapeutic potential of targeting the BRAF in patients with colorectal cancer. J. Cell Physiol. 2017, 9999, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Shi, Y.L.; Zhou, K.; Wang, L.L.; Yan, Z.X.; Liu, Y.L.; Xu, L.L.; Zhao, S.W.; Chu, H.L.; Shi, T.T.; et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018, 9, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Di Nicolantonio, F.; Martini, M.; Molinari, F.; Sartore-Bianchi, A.; Arena, S.; Saletti, P.; De Dosso, S.; Mazzucchelli, L.; Frattini, M.; Siena, S.; et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 5705–5712. [Google Scholar] [CrossRef] [PubMed]
- Cappuzzo, F.; Varella-Garcia, M.; Finocchiaro, G.; Skokan, M.; Gajapathy, S.; Carnaghi, C.; Rimassa, L.; Rossi, E.; Ligorio, C.; Tommaso, L.D. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br. J. Cancer 2008, 99, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Affolter, K.; Samowitz, W.; Tripp, S.; Bronner, M.P. BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes Chromosomes Cancer 2013, 52, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Benlloch, S.; Payá, A.; Alenda, C.; Bessa, X.; Andreu, M.; Jover, R.; Castells, A.; Llor, X.; Aranda, F.L. Detection of BRAF V600E mutation in colorectal cancer: Comparison of automatic sequencing and real-time chemistry methodology. J. Mol. Diagn. 2006, 8, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.H.; Liu, Y.; Eu, K.W.; Ang, P.W.; Li, W.Q.; Salto-Tellez, M.; Iacopetta, B.; Soong, R. Detection of BRAF V600E mutation by pyrosequencing. Pathology 2008, 40, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Lavine, B.; Workman, J. Chemometrics. Anal. Chem. 2010, 82, 4699–4711. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Martín, E.M.; García-García, M.D.C.; Font, R.; Moreno-Rojas, J.M.; Salinas-Navarro, M.; Gómez, P.; Río-Celestino, M.D. Quantification of total phenolic and carotenoid content in blackberries (Rubus fructicosus L.) using near infrared spectroscopy (NIRS) and multivariate analysis. Molecules 2018, 23, 3191. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Li, C.; Zhao, N.; Li, H.; Chang, Q.; Liu, X.; Liao, Y.; Pan, R. Rapid determination of active compounds and antioxidant activity of okra seeds using fourier transform near infrared (FT-NIR) spectroscopy. Molecules 2018, 23, 550. [Google Scholar] [CrossRef]
- Nioka, S.; Chance, B. NIR spectroscopic detection of breast cancer. Technol. Cancer Res. Treat. 2005, 4, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Tian, J.; Xiang, Y.; Zhang, Z.; Harrington, P.D.B. Near infrared spectroscopy combined with least squares support vector machines and fuzzy rule-building expert system applied to diagnosis of endometrial carcinoma. Cancer Epidemiol. 2012, 36, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Cui, D.; Li, Z.; Wu, L.; Shen, A.; Hu, J. Gastric cancer differentiation using fourier transform near-infrared spectroscopy with unsupervised pattern recognition. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2013, 101, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lin, Z.; Wu, H.; Wang, L.; Wu, T.; Tan, C. Diagnosis of colorectal cancer by near-infrared optical fiber spectroscopy and random forest. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 135, 185–191. [Google Scholar] [CrossRef] [PubMed]
- McClure, W.F. 204 years of near infrared technology: 1800–2003. J. Near Infrared Spectrosc. 2003, 11, 487–518. [Google Scholar] [CrossRef]
- Pasquini, C. Near infrared spectroscopy: A mature analytical technique with new perspectives—A review. Anal. Chim. Acta 2018, 1026, 8–36. [Google Scholar] [CrossRef] [PubMed]
- Workman, J.; Weyer, L. Practical Guide to Interpretive Nearinfrared Spectroscopy; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Lu, W.Z. Modern near Infrared Spectroscopy Analytical Technology, 2nd ed.; China Petrochemical Press: Beijing, China, 2006; pp. 46–47. [Google Scholar]
- Fan, Q.; Cao, L.Y.; Wang, Y.L.; Chen, Y.; Dong, Y.H. A Fast Identification Method of Human Colorectal Cancer Tissue by near Infrared Diffuse Reflectance Spectroscopy: 201410353552.0. 2018-03-30. Available online: http://epub.cnipa.gov.cn/patentoutline.action (accessed on 9 May 2019).
Sample Availability: Not available. |




| Model Number | Class of Samples | Number of Calibration Samples | Number of Validation Samples | ||
|---|---|---|---|---|---|
| Mutant | Wild-type | Mutant | Wild-type | ||
| 1 | Class 1 | 40 | 40 | 12 | 12 |
| 2 | Class 2 | 40 | 40 | 12 | 12 |
| 3 | Class 3 | 40 | 40 | 12 | 12 |
| 4 | Class 2&1 | 20&20 | 20&20 | NA | NA |
| 5 | Class 2&3 | 20&20 | 20&20 | NA | NA |
| Model Number | Preprocessing | Spectral Subrange (cm−1) | Number of PCs/ Cumulative Variance Contribution Rate (%) | Number of Neurons on Each Side | Model Performances | ||
|---|---|---|---|---|---|---|---|
| CAC (%) | CACV (%) | CAV (%) | |||||
| 1 | MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 98.0 | 95.0 | 94.4 |
| 1.1 | MSC + MC | 9000–6800, 6500–4000 | 6/99.9 | 12 | 97.0 | 93.0 | 90.3 |
| 1.2 | SNV + MC | 9000–6800, 6500–4000 | 6/99.9 | 12 | 97.0 | 94.0 | 81.9 |
| 1.3 | FD + MC | 9000–6800, 6500–4000 | 6/98.8 | 12 | 93.0 | 86.0 | 88.9 |
| 1.4 | SD + MC | 9000–6800, 6500–4000 | 6/95.7 | 12 | 89.0 | 71.0 | 73.6 |
| 1.5 | SGS + MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 98.0 | 94.0 | 90.3 |
| 1.6 | SGS + FD + MC | 9000–6800, 6500–4000 | 6/99.1 | 12 | 94.0 | 88.0 | 90.3 |
| 1.7 | NDS + FD + MC | 9000–6800, 6500–4000 | 3/100.0 | 12 | 92.0 | 85.0 | 87.5 |
| 1.8 | MSC + SD + MC | 9000–6800, 6500–4000 | 6/ 96.0 | 12 | 90.0 | 74.0 | 77.8 |
| 1.9 | SNV + NDS + FD + MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 95.0 | 88.0 | 90.3 |
| 1.10 | MC | 9000–4000 | 6/100.0 | 12 | 98.0 | 94.0 | 91.7 |
| 1.11 | MC | 9000–6800, 6500–4000 | 6/100.0 | 10 | 97.0 | 94.0 | 88.9 |
| 1.12 | MC | 9000–6800, 6500–4000 | 6/100.0 | 15 | 98.0 | 96.0 | 88.9 |
| 2 | MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 97.0 | 92.0 | 94.4 |
| 2.1 | MSC + MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 94.0 | 85.0 | 79.2 |
| 2.2 | SNV + MC | 9000–6800, 6500–4000 | 6/ 99.9 | 12 | 89.0 | 83.0 | 83.3 |
| 2.3 | FD + MC | 9000–6800, 6500–4000 | 6/97.2 | 12 | 90.0 | 82.0 | 86.1 |
| 2.4 | SD + MC | 9000–6800, 6500–4000 | 20/84.6 | 12 | NA | NA | NA |
| 2.5 | SGS + MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 96.0 | 94.0 | 90.3 |
| 2.6 | SGS + FD + MC | 9000–6800, 6500–4000 | 6/97.8 | 12 | 92.0 | 88.0 | 81.9 |
| 2.7 | NDS + FD + MC | 9000–6800, 6500–4000 | 2/100.0 | 12 | 88.0 | 80.0 | 79.2 |
| 2.8 | MSC + SD + MC | 9000–6800, 6500–4000 | 20/80.6 | 12 | NA | NA | NA |
| 2.9 | SNV + NDS + FD + MC | 9000–6800, 6500–4000 | 3/100.0 | 12 | 90.0 | 85.0 | 87.5 |
| 2.10 | MC | 9000–4000 | 6/100.0 | 12 | 96.0 | 91.0 | 93.1 |
| 2.11 | MC | 9000–6800, 6500–4000 | 6/100.0 | 10 | 96.0 | 90.0 | 87.5 |
| 2.12 | MC | 9000–6800, 6500–4000 | 6/100.0 | 15 | 97.0 | 92.0 | 94.4 |
| 3 | MC | 9000–6800, 6500–4000 | 5/100.0 | 12 | 95.0 | 88.0 | 93.1 |
| 3.1 | MSC + MC | 9000–6800, 6500–4000 | 5/99.9 | 12 | 86.0 | 71.0 | 66.7 |
| 3.2 | SNV + MC | 9000–6800, 6500–4000 | 5/99.9 | 12 | 85.0 | 72.0 | 68.1 |
| 3.3 | FD + MC | 9000–6800, 6500–4000 | 13/85.5 | 12 | 90.0 | 77.0 | 79.2 |
| 3.4 | SD + MC | 9000–6800, 6500–4000 | 20/75.0 | 12 | NA | NA | NA |
| 3.5 | SGS + MC | 9000–6800, 6500–4000 | 5/100.0 | 12 | 93.0 | 89.0 | 90.3 |
| 3.6 | SGS + FD + MC | 9000–6800, 6500–4000 | 10/ 85.6 | 12 | 88.0 | 79.0 | 76.4 |
| 3.7 | NDS + FD + MC | 9000–6800, 6500–4000 | 2/100.0 | 12 | 90.0 | 82.0 | 77.8 |
| 3.8 | MSC + SD + MC | 9000–6800, 6500–4000 | 20/74.5 | 12 | NA | NA | NA |
| 3.9 | SNV + NDS + FD + MC | 9000–6800, 6500–4000 | 4/100.0 | 12 | 87.0 | 65.0 | 72.2 |
| 3.10 | MC | 9000–4000 | 5/100.0 | 12 | 95.0 | 88.0 | 87.5 |
| 3.11 | MC | 9000–6800, 6500–4000 | 5/100.0 | 10 | 93.0 | 89.0 | 86.1 |
| 3.12 | MC | 9000–6800, 6500–4000 | 5/100.0 | 15 | 95.0 | 89.0 | 88.9 |
| 4 | MC | 9000–6800, 6500–4000 | 5/100.0 | 12 | 97.0 | 97.0 | NA |
| 5 | MC | 9000–6800, 6500–4000 | 6/100.0 | 12 | 95.0 | 90.0 | NA |
| Model Number | Diagnostic Performances | ||
|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Accuracy (%) | |
| 1 | 100.0 | 87.5 | 93.8 |
| 2 | 100.0 | 95.0 | 97.5 |
| 3 | 100.0 | 82.5 | 91.3 |
| 4 | 100.0 | 92.5 | 96.3 |
| 5 | 100.0 | 85.0 | 92.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yang, Y.; Wang, Y.; Fan, Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules 2019, 24, 2238. https://doi.org/10.3390/molecules24122238
Zhang X, Yang Y, Wang Y, Fan Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules. 2019; 24(12):2238. https://doi.org/10.3390/molecules24122238
Chicago/Turabian StyleZhang, Xue, Yang Yang, Yalan Wang, and Qi Fan. 2019. "Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network" Molecules 24, no. 12: 2238. https://doi.org/10.3390/molecules24122238