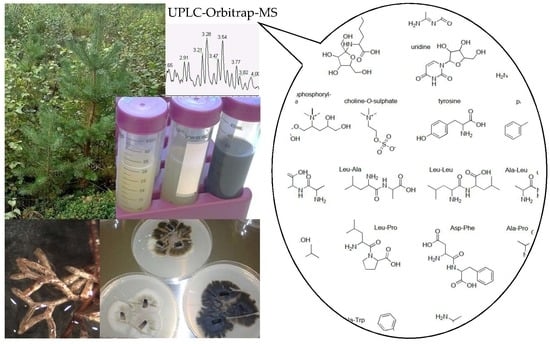
Metabolic Profiling of Water-Soluble Compounds from the Extracts of Dark Septate Endophytic Fungi (DSE) Isolated from Scots Pine (Pinus sylvestris L.) Seedlings Using UPLC–Orbitrap–MS
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of the Endophytic Fungal Isolates
2.2. Identification of the Metabolites
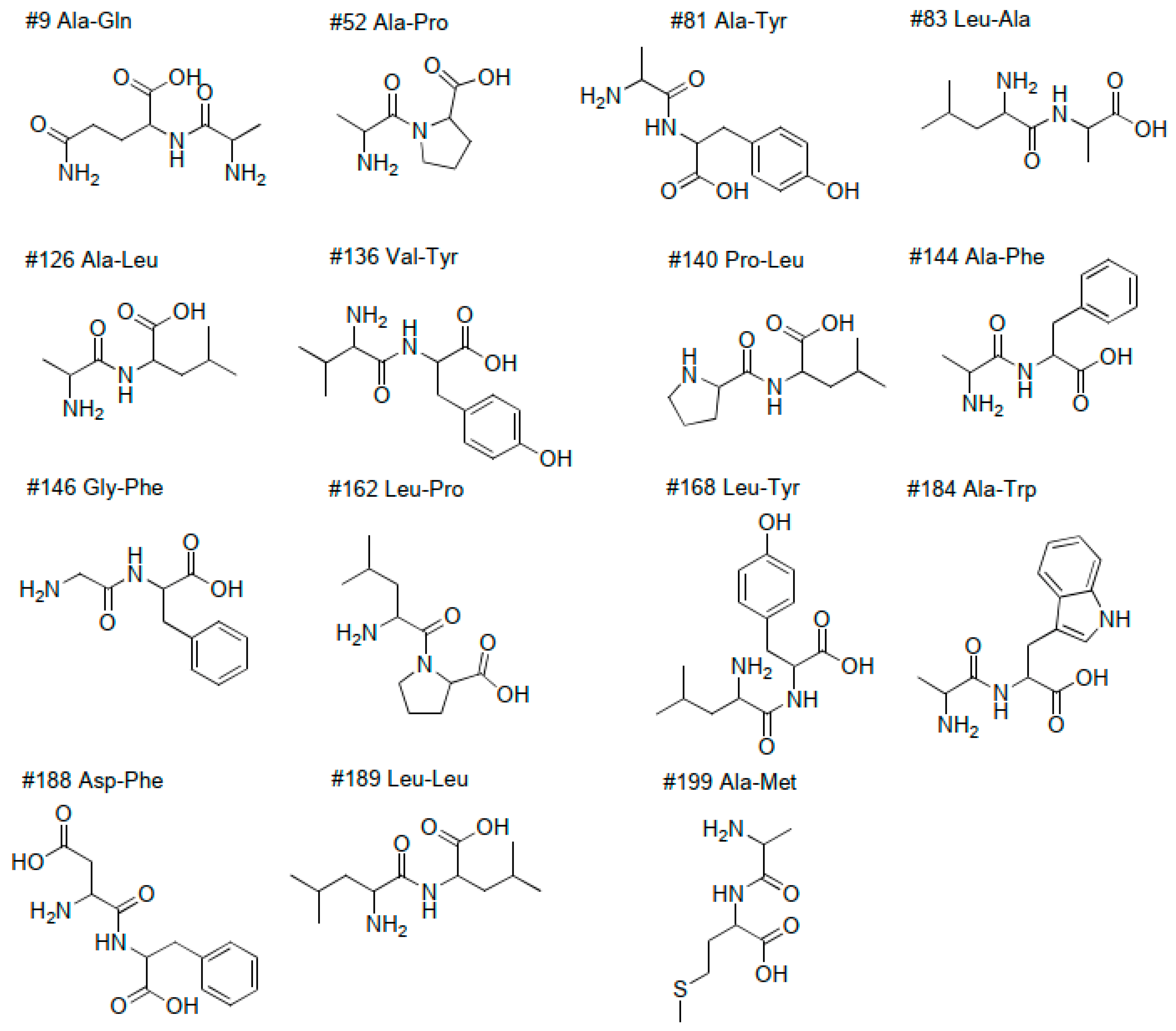
2.2.1. Amino Acids, Dipeptides and Peptides
2.2.2. Opine Amino Acids
2.2.3. Amino Acid Quinones and Amadori Compounds
2.2.4. Cholines
2.2.5. Nucleobases, Nucleosides, Nucleotides, and their Derivatives
2.2.6. Siderophores
2.2.7. Other Common Metabolites
2.2.8. Sugars, Sugar Alcohols, Disaccharides
2.2.9. Endophyte or Plant Metabolites
2.3. Metabolites in Fungal Extracts
3. Materials and Methods
3.1. Reagents
3.2. Endophytic Fungi Isolation and Identification
3.3. Fungal Extract Preparation
3.4. UHPLC-DAD-ESI-Orbitrap-MS
3.5. Identification
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stone, J.K.; Bacon, C.W.; White, J. An Overview of Endophytic Microbes: Endophytism defined. In Microbial Endophytes, 1st ed.; Bacon, C.W., White, J.F., Jr., Eds.; Elsevier Inc.: New York, NY, USA, 2000; pp. 17–24. [Google Scholar]
- Schulz, B.; Boyle, C. What are Endophytes? In Microbial Root Endophytes, 1st ed.; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer Science & Business Media: Berlin, Germany, 2006; pp. 1–13. [Google Scholar]
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- Strobel, G. The Emergence of Endophytic Microbes and Their Biological Promise. J. Fungi 2018, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Waller, F.; Achatz, B.; Baltruschat, H.; Fodor, J.; Becker, K.; Fischer, M.; Heier, T.; Huckelhoven, R.; Neumann, C.; Von Wettstein, D.; et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. USA 2005, 102, 13386–13391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.W.; Song, Y.C.; Tan, R.X. Biology and chemistry of endophytes. Nat. Prod. Rep. 2006, 23, 753–771. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Nagabhyru, P.; Dinkins, R.D.; Wood, C.L.; Bacon, C.W.; Schardl, C.L. Tall fescue endophyte effects on tolerance to water-deficit stress. BMC Plant Boil. 2013, 13, 127. [Google Scholar] [CrossRef] [PubMed]
- Tellenbach, C.; Sumarah, M.W.; Grünig, C.R.; Miller, J.D. Inhibition of Phytophthora species by secondary metabolites produced by the dark septate endophyte Phialocephala europaea. Fungal Ecol. 2013, 6, 12–18. [Google Scholar] [CrossRef]
- Terhonen, E.; Keriö, S.; Sun, H.; Asiegbu, F.O. Endophytic fungi of Norway spruce roots in boreal pristine mire, drained peatland and mineral soil and their inhibitory effect on Heterobasidion parviporum in vitro. Fungal Ecol. 2014, 9, 17–26. [Google Scholar] [CrossRef]
- Grünig, C.R.; Queloz, V.; Sieber, T.N.; Holdenrieder, O. Dark septate endophytes (DSE) of the Phialocephala fortinii s.l. –Acephala applanata species complex in tree roots: Classification, population biology, and ecology. Botany 2008, 86, 1355–1369. [Google Scholar] [CrossRef]
- Jumpponen, A.; Trappe, J.M. Dark septate endophytes: A review of facultative biotrophic root-colonizing fungi. New Phytol. 1998, 140, 295–310. [Google Scholar] [CrossRef]
- Mandyam, K.; Jumpponen, A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud. Mycol. 2005, 53, 173–189. [Google Scholar] [CrossRef]
- Schulz, B. Mutualistic Interactions with Fungal Root Endophytes. In Microbial Root Endophytes, 1st ed.; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer Science & Business Media: Berlin, Germany, 2006; pp. 261–279. [Google Scholar]
- Tellenbach, C.; Grünig, C.R.; Sieber, T.N. Negative effects on survival and performance of Norway spruce seedlings colonized by dark septate root endophytes are primarily isolate-dependent. Environ. Microbiol. 2011, 13, 2508–2517. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, R.M.; Gonzalez, A.M.; Ramirez, A.M. Compounds Derived from Endophytes: A Review of Phytochemistry and Pharmacology. Curr. Med. Chem. 2012, 19, 2992–3030. [Google Scholar] [CrossRef] [PubMed]
- Koskimäki, J.J.; Kajula, M.; Hokkanen, J.; Ihantola, E.-L.; Kim, J.H.; Hautajärvi, H.; Hankala, E.; Suokas, M.; Pohjanen, J.; Podolich, O.; et al. Methyl-esterified 3-hydroxybutyrate oligomers protect bacteria from hydroxyl radicals. Nat. Chem. Biol. 2016, 12, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Shashidhar, M.; Giridhar, P.; Sankar, K.U.; Manohar, B. Bioactive principles from Cordyceps sinensis: A potent food supplement—A review. J. Funct. Foods 2013, 5, 1013–1030. [Google Scholar] [CrossRef]
- Endo, N.; Dokmai, P.; Suwannasai, N.; Phosri, C.; Horimai, Y.; Hirai, N.; Fukuda, M.; Yamada, A. Ectomycorrhization of Tricholoma matsutake with Abies veitchii and Tsuga diversifolia in the subalpine forests of Japan. Mycoscience 2015, 56, 402–412. [Google Scholar] [CrossRef]
- Khan, Z.; Gené, J.; Ahmad, S.; Cano, J.; Al-Sweih, N.; Joseph, L.; Chandy, R.; Guarro, J. Coniochaeta polymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecythophora species to Coniochaeta. Antonie van Leeuwenhoek 2013, 104, 243–252. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef]
- Meek, J.L. Prediction of peptide retention times in high-pressure liquid chromatography on the basis of amino acid composition. Proc. Natl. Acad. Sci. USA 1980, 77, 1632–1636. [Google Scholar] [CrossRef]
- Liu, Z.; Rochfort, S. A fast liquid chromatography–mass spectrometry (LC–MS) method for quantification of major polar metabolites in plants. J. Chromatogr. B 2013, 912, 8–15. [Google Scholar] [CrossRef]
- Liang, K.; Liang, S.; Lu, L.; Zhu, D.; Zhu, H.; Liu, P.; Zhang, M. Metabolic variation and cooking qualities of millet cultivars grown both organically and conventionally. Food Res. Int. 2018, 106, 825–833. [Google Scholar] [CrossRef]
- Sun, L.-M.; Zhang, B.; Wang, Y.-C.; He, H.-K.; Chen, X.-G.; Wang, S.-J. Metabolomic analysis of raw Pinelliae Rhizoma and its alum-processed products via UPLC-MS and their cytotoxicity. Biomed. Chromatogr. 2019, 33, e4411. [Google Scholar] [CrossRef]
- Ryu, K.; Ide, N.; Matsuura, H.; Itakura, Y. Nα-(1-Deoxy-D-fructos-1-yl)-L-Arginine, an Antioxidant Compound Identified in Aged Garlic Extract. J. Nutr. 2001, 131, 972S–976S. [Google Scholar] [CrossRef]
- Yoshida, N.; Takatsuka, K.; Katsuragi, T.; Tani, Y. Occurrence of Fructosyl-Amino Acid Oxidase-Reactive Compounds in Fungal Cells. Biosci. Biotechnol. Biochem. 2005, 69, 258–260. [Google Scholar] [CrossRef]
- Wang, J.; Lu, Y.-M.; Liu, B.-Z.; He, H.-Y. Electrospray positive ionization tandem mass spectrometry of Amadori compounds. J. Mass Spectrom. 2008, 43, 262–264. [Google Scholar] [CrossRef]
- Prior, S.L.; Cunliffe, B.W.; Robson, G.D.; Trinci, A.P. Multiple isomers of phosphatidyl inositol monophosphate and inositol bis- and trisphosphates from filamentous fungi. FEMS Microbiol. Lett. 1993, 110, 147–152. [Google Scholar] [CrossRef]
- Van Der Rest, B.; Boisson, A.-M.; Gout, E.; Bligny, R.; Douce, R. Glycerophosphocholine Metabolism in Higher Plant Cells. Evidence of a New Glyceryl-Phosphodiester Phosphodiesterase. Plant Physiol. 2002, 130, 244–255. [Google Scholar] [CrossRef]
- Landi, N.; Pacifico, S.; Ragucci, S.; Di Giuseppe, A.M.; Iannuzzi, F.; Zarrelli, A.; Piccolella, S.; Di Maro, A. Pioppino mushroom in southern Italy: An undervalued source of nutrients and bioactive compounds. J. Sci. Food Agric. 2017, 97, 5388–5397. [Google Scholar] [CrossRef]
- Gamal-Eldin, M.A.; Macartney, D.H. Selective molecular recognition of methylated lysines and arginines by cucurbit[6]uril and cucurbit[7]uril in aqueous solution. Org. Biomol. Chem. 2013, 11, 488–495. [Google Scholar] [CrossRef]
- Chilton, W.S.; Petit, A.; Chilton, M.-D.; Dessaux, Y. Structure and characterization of the crown gall opines heliopine, vitopine and ridéopine. Phytochem. 2001, 58, 137–142. [Google Scholar] [CrossRef]
- Shiao, M.-S.; Wang, Z.-N.; Lin, L.-J.; Lien, J.-Y.; Wang, J.-J. Profiles of nucleosides and nitrogen bases in Chinese medicinal fungus Cordyceps sinensis and related species. Bot. Bull. Acad. Sin. 1994, 35, 261–267. [Google Scholar]
- Ranogajec, A.; Beluhan, S.; Šmit, Z. Analysis of nucleosides and monophosphate nucleotides from mushrooms with reversed-phase HPLC. J. Sep. Sci. 2010, 33, 1024–1033. [Google Scholar] [CrossRef]
- Dudley, E.; Bond, L. Mass spectrometry analysis of nucleosides and nucleotides. Mass Spectrom. Rev. 2014, 33, 302–331. [Google Scholar] [CrossRef]
- Laourdakis, C.D.; Merino, E.F.; Neilson, A.P.; Cassera, M.B. Comprehensive quantitative analysis of purines and pyrimidines in the human malaria parasite using ion-pairing ultra-performance liquid chromatography-mass spectrometry. J. Chromatogr. B 2014, 967, 127–133. [Google Scholar] [CrossRef]
- Li, S.; Jin, Y.; Tang, Z.; Lin, S.; Liu, H.; Jiang, Y.; Cai, Z. A novel method of liquid chromatography–tandem mass spectrometry combined with chemical derivatization for the determination of ribonucleosides in urine. Anal. Chim. Acta 2015, 864, 30–38. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, Q.; Wang, M.; Fu, S.; Zhang, Q.; Zhang, Z.; Zhao, H.; Liu, Y.; Huang, Z.; Xie, Z.; et al. Using UHPLC Q-Trap/MS as a complementary technique to in-depth mine UPLC Q-TOF/MS data for identifying modified nucleosides in urine. J. Chromatogr. B 2017, 1051, 108–117. [Google Scholar] [CrossRef]
- Markham, P.; Robson, G.D.; Bainbridge, B.W.; Trinci, A.P. Choline: Its role in the growth of filamentous fungi and the regulation of mycelial morphology. FEMS Microbiol. Lett. 1993, 104, 287–300. [Google Scholar] [CrossRef]
- Lamberts, L.; Rombouts, I.; Delcour, J.A. Study of nonenzymic browning in α-amino acid and γ-aminobutyric acid/sugar model systems. Food Chem. 2008, 111, 738–744. [Google Scholar] [CrossRef]
- Kemp, J.D. In Vivo Synthesis of Crown Gall-specific Agrobacterium tumefaciens-directed Derivatives of Basic Amino Acids. Plant Physiol. 1978, 62, 26–30. [Google Scholar] [CrossRef]
- Spencer, B.; Hussey, E.C.; Orsi, B.A.; Scott, J.M. Mechanism of choline O-sulphate utilization in fungi. Biochem. J. 1968, 106, 461–469. [Google Scholar] [CrossRef]
- Grass, J.; Pabst, M.; Kolarich, D.; Pöltl, G.; Léonard, R.; Brecker, L.; Altmann, F. Discovery and Structural Characterization of Fucosylated Oligomannosidic N-Glycans in Mushrooms. J. Boil. Chem. 2010, 286, 5977–5984. [Google Scholar] [CrossRef]
- Kariya, M.; Namiki, H. HPLC of phenylthiocarbamyol labelled uridine-diphosphate-hexosamine. Chromatographia 1997, 46, 5–11. [Google Scholar] [CrossRef]
- El-Ganiny, A.M.; Sheoran, I.; Sanders, D.A.; Kaminskyj, S.G. Aspergillus nidulans UDP-glucose-4-epimerase UgeA has multiple roles in wall architecture, hyphal morphogenesis, and asexual development. Fungal Genet. Boil. 2010, 47, 629–635. [Google Scholar] [CrossRef]
- Carpi, F.M.; Cortese, M.; Orsomando, G.; Polzonetti, V.; Vincenzetti, S.; Moreschini, B.; Coleman, M.; Magni, G.; Pucciarelli, S. Simultaneous quantification of nicotinamide mononucleotide and related pyridine compounds in mouse tissues by UHPLC-MS/MS. Sep. Sci. PLUS 2018, 1, 22–30. [Google Scholar] [CrossRef]
- Maruyama, D.; Nishitani, Y.; Nonaka, T.; Kita, A.; Fukami, T.A.; Mio, T.; Yamada-Okabe, H.; Yamada-Okabe, T.; Miki, K. Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism. J. Boil. Chem. 2007, 282, 17221–17230. [Google Scholar] [CrossRef]
- Strijbis, K.; Distel, B. Intracellular Acetyl Unit Transport in Fungal Carbon Metabolism. Eukaryot. Cell 2010, 9, 1809–1815. [Google Scholar] [CrossRef]
- Lorenzetti, R.; Lilla, S.; Donato, J.L.; De Nucci, G. Simultaneous quantification of GMP, AMP, cyclic GMP and cyclic AMP by liquid chromatography coupled to tandem mass spectrometry. J. Chromatogr. B 2007, 859, 37–41. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, S.; Paintsil, E.; Dutschman, G.E.; Gullen, E.A.; Chu, E.; Cheng, Y.-C. Analysis of deoxyribonucleotide pools in human cancer cell lines using a liquid chromatography coupled with tandem mass spectrometry technique. Biochem. Pharmacol. 2011, 82, 411–417. [Google Scholar] [CrossRef]
- Zhu, M.; Assmann, S.M. Metabolic Signatures in Response to Abscisic Acid (ABA) Treatment in Brassica napus Guard Cells Revealed by Metabolomics. Sci. Rep. 2017, 7, 12875. [Google Scholar] [CrossRef]
- Li, M.; Chen, T.; Gao, T.; Miao, Z.; Jiang, A.; Shi, L.; Ren, A.; Zhao, M. UDP-glucose pyrophosphorylase influences polysaccharide synthesis, cell wall components, and hyphal branching in Ganoderma lucidum via regulation of the balance between glucose-1-phosphate and UDP-glucose. Fungal Genet. Boil. 2015, 82, 251–263. [Google Scholar] [CrossRef]
- Ibanez, C.; Simó, C.; Garcia-Cañas, V.; Gómez-Martínez, Á.; Ferragut, J.A.; Cifuentes, A. CE/LC-MS multiplatform for broad metabolomic analysis of dietary polyphenols effect on colon cancer cells proliferation. Electrophoresis 2012, 33, 2328–2336. [Google Scholar] [CrossRef]
- Bogo, A.; Mantle, P.G.; Casa, R.T.; Guidolin, A.F. The structures of the honeydew oligosaccharides synthesized by Claviceps africana. Summa Phytopathol. 2006, 32, 16–20. [Google Scholar] [CrossRef]
- Miyakoshi, S.; Uchiyama, H.; Someya, T.; Satoh, T.; Tabuchi, T. Distribution of the Methylcitric Acid Cycle and ^-Oxidation Pathway for Propionate Catabolism in Fungi. Agric. Boil. Chem. 1987, 51, 2381–2387. [Google Scholar] [CrossRef]
- Brock, M.; Maerker, C.; Schutz, A.; Völker, U.; Buckel, W. Oxidation of propionate to pyruvate in Escherichia coli: Involvement of methylcitrate dehydratase and aconitase. Eur. J. Biochem. 2002, 269, 6184–6194. [Google Scholar] [CrossRef]
- Bähre, H.; Kaever, V. Measurement of 2′,3′-cyclic nucleotides by liquid chromatography–tandem mass spectrometry in cells. J. Chromatogr. B 2014, 964, 208–211. [Google Scholar] [CrossRef]
- Dittmar, F.; Abdelilah-Seyfried, S.; Tschirner, S.K.; Kaever, V.; Seifert, R. Temporal and organ-specific detection of cNMPs including cUMP in the zebrafish. Biochem. Biophys. Res. Commun. 2015, 468, 708–712. [Google Scholar] [CrossRef]
- Seifert, R. Distinct Signaling Roles of cIMP, cCMP, and cUMP. Structure 2016, 24, 1627–1628. [Google Scholar] [CrossRef]
- Świeżawska, B.; Duszyn, M.; Jaworski, K.; Szmidt-Jaworska, A. Downstream Targets of Cyclic Nucleotides in Plants. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef]
- Contreras-Sanz, A.; Scott-Ward, T.S.; Gill, H.S.; Jacoby, J.C.; Birch, R.E.; Malone-Lee, J.; Taylor, K.M.G.; Peppiatt-Wildman, C.M.; Wildman, S.S.P. Simultaneous quantification of 12 different nucleotides and nucleosides released from renal epithelium and in human urine samples using ion-pair reversed-phase HPLC. Purinergic Signal. 2012, 8, 741–751. [Google Scholar] [CrossRef]
- Perret, D. Of urines and purines—A life in separation science. Nucleosides Nucleotides Nucleic Acids 2018, 37, 588–602. [Google Scholar] [CrossRef]
- Pocsi, I.; Prade, R.A.; Penninckx, M.J. Glutathione, Altruistic metabolite in fungi. Adv. Microb. Physiol. 2004, 49, 1–76. [Google Scholar] [CrossRef]
- Semighini, C.P.; Savoldi, M.; Goldman, G.H.; Harris, S.D. Functional Characterization of the Putative Aspergillus nidulans Poly(ADP-Ribose) Polymerase Homolog PrpA. Genetics 2006, 173, 87–98. [Google Scholar] [CrossRef]
- Farag, M.A.; El-Kersh, D.M.; Ehrlich, A.; Choucry, M.A.; El-Seedi, H.; Frolov, A.; Wessjohann, L.A.; Shokry, M.; Frolov, A. Variation in Ceratonia siliqua pod metabolome in context of its different geographical origin, ripening stage and roasting process. Food Chem. 2019, 283, 675–687. [Google Scholar] [CrossRef]
- Giffhorn, F. Fungal pyranose oxidases: Occurrence, properties and biotechnical applications in carbohydrate chemistry. Appl. Microbiol. Biotechnol. 2000, 54, 727–740. [Google Scholar] [CrossRef]
- Nain-Perez, A.; Barbosa, L.C.A.; Maltha, C.R.Á.; Forlani, G. Natural Abenquines and Their Synthetic Analogues Exert Algicidal Activity against Bloom-Forming Cyanobacteria. J. Nat. Prod. 2017, 80, 813–818. [Google Scholar] [CrossRef]
- Schulz, D.; Beese, P.; Ohlendorf, B.; Erhard, A.; Zinecker, H.; Dorador, C.; Imhoff, J.F. Abenquines A–D: aminoquinone derivatives produced by Streptomyces sp. strain DB634. J. Antibiot. 2011, 64, 763–768. [Google Scholar] [CrossRef]
- Cheng, Q.; Gu, J.; Compaan, K.R.; Schaefer, H.F. Isoguanine Formation from Adenine. Chem. - A Eur. J. 2012, 18, 4877–4886. [Google Scholar] [CrossRef]
- Karalkar, N.B.; Khare, K.; Molt, R.; Benner, S.A. Tautomeric equilibria of isoguanine and related purine analogs. Nucleosides Nucleotides Nucleic Acids 2017, 31, 1–19. [Google Scholar] [CrossRef]
- Bartholdy, B.; Berreck, M.; Haselwandter, K. Hydroxamate siderophore synthesis by Phialocephala fortinii, a typical dark septate fungal root endophyte. BioMetals 2001, 14, 33–42. [Google Scholar] [CrossRef]
- Haas, H.; Eisendle, M.; Turgeon, B.G. Siderophores in Fungal Physiology and Virulence. Annu. Rev. Phytopathol. 2008, 46, 149–187. [Google Scholar] [CrossRef]
- Holinsworth, B.; Martin, J.D. Siderophore production by marine-derived fungi. BioMetals 2009, 22, 625–632. [Google Scholar] [CrossRef]
- Bertrand, S.; Duval, O.; Helesbeux, J.-J.; Larcher, G.; Richomme, P. Synthesis of the trans-fusarinine scaffold. Tetrahedron Lett. 2010, 51, 2119–2122. [Google Scholar] [CrossRef]
- Kajula, M.; Tejesvi, M.V.; Kolehmainen, S.; Mäkinen, A.; Hokkanen, J.; Mattila, S.; Pirttilä, A.-M. The siderophore ferricrocin produced by specific foliar endophytic fungi in vitro. Fungal Biol. 2010, 114, 248–254. [Google Scholar] [CrossRef]
- Rublee, P.A.; Merkel, S.M.; Faust, M.A.; Miklas, J. Distribution and activity of bacteria in the headwaters of the Rhode River Estuary, Maryland, USA. Microb. Ecol. 1984, 10, 243–255. [Google Scholar] [CrossRef]
- Klinger, M.M.; A Laine, R.; Steiner, S.M. Characterization of novel amino acid fucosides. J. Boil. Chem. 1981, 256, 7932–7935. [Google Scholar]
- Cuendet, M.; Hostettmann, K.; Potterat, O.; Dyatmiko, W. Iridoid Glucosides with Free Radical Scavenging Properties from Fagraea blumei. Helvetica Chim. Acta 1997, 80, 1144–1152. [Google Scholar] [CrossRef]
- Hill, R.A.; Sutherland, A. Hot off the press. Nat. Prod. Rep. 2017, 34, 338–342. [Google Scholar] [CrossRef]
- Sang, X.-N.; Chen, S.-F.; Chen, G.; An, X.; Li, S.-G.; Lu, X.-J.; Zhao, D.; Bai, J.; Wang, H.-F.; Pei, Y.-H. Two pairs of enantiomeric α-pyrone dimers from the endophytic fungus Phoma sp. YN02-P-3. RSC Adv. 2017, 7, 1943–1946. [Google Scholar] [CrossRef]
- Wang, X.; Lin, M.; Xu, D.; Lai, D.; Zhou, L. Structural Diversity and Biological Activities of Fungal Cyclic Peptides, Excluding Cyclodipeptides. Molecules 2017, 22, 2069. [Google Scholar] [CrossRef]
- Li, X.-Y.; Wang, Y.-H.; Yang, J.; Cui, W.-Y.; He, P.-J.; Munir, S.; He, P.-F.; Wu, Y.-X.; He, Y.-Q. Acaricidal Activity of Cyclodipeptides from Bacillus amyloliquefaciens W1 against Tetranychus urticae. J. Agric. Food Chem. 2018, 66, 10163–10168. [Google Scholar] [CrossRef]
- Yamaoka, N.; Kudo, Y.; Inazawa, K.; Inagawa, S.; Yasuda, M.; Mawatari, K.-I.; Nakagomi, K.; Kaneko, K. Simultaneous determination of nucleosides and nucleotides in dietary foods and beverages using ion-pairing liquid chromatography–electrospray ionization-mass spectrometry. J. Chromatogr. B 2010, 878, 2054–2060. [Google Scholar] [CrossRef]
- Sánchez, J.; Nikolau, B.J.; Stumpf, P.K. Reduction of N-Acetyl Methionine Sulfoxide in Plants. Plant Physiol. 1983, 73, 619–623. [Google Scholar] [CrossRef]
- Ling, S.-K.; Komorita, A.; Tanaka, T.; Fujioka, T.; Mihashi, K.; Kouno, I. Iridoids and Anthraquinones from the Malaysian Medicinal Plant, Saprosma scortechinii (Rubiaceae). Chem. Pharm. Bull. 2002, 50, 1035–1040. [Google Scholar] [CrossRef]
- Kanchanapoom, T.; Kasai, R.; Yamasaki, K. Iridoid and phenolic glycosides from Morinda coreia. Phytochemistry 2002, 59, 551–556. [Google Scholar] [CrossRef]
- Li, C.; Xue, X.; Zhou, D.; Zhang, F.; Xu, Q.; Ren, L.; Liang, X. Analysis of iridoid glucosides in Hedyotis diffusa by high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 2008, 48, 205–211. [Google Scholar] [CrossRef]
- Friščić, M.; Bucar, F.; Pilepić, K.H. LC-PDA-ESI-MS analysis of phenolic and iridoid compounds from Globularia spp. J. Mass Spectrom. 2016, 51, 1211–1236. [Google Scholar] [CrossRef]
- Alara, O.R.; Abdurahman, N.H.; Ukaegbu, C.I.; Azhari, N.H.; Kabbashi, N.A. Metabolic profiling of flavonoids, saponins, alkaloids, and terpenoids in the extract from Vernonia cinerea leaf using LC-Q-TOF-MS. J. Liq. Chromatogr. Relat. Technol. 2018, 41, 722–731. [Google Scholar] [CrossRef]
- Fushiya, S.; Matsuda, M.; Yamada, S.; Nozoe, S. New opine type amino acids from a poisonous mushroom, Clitocybe acromelalga. Tetrahedron 1996, 52, 877–886. [Google Scholar] [CrossRef]
- Davidek, T.; Blank, I.; Kraehenbuehl, K.; Hau, J.; Devaud, S. New approaches in the analysis of Amadori compounds. In Innovations in Analytical Flavor Research, State-of-the-Art in Flavour Chemistry and Biology, Proceedings of the 7th Wartburg Symposium, Eisenach, Germany, 21–23 April 2004; Hofmann, T., Rothe, M., Schieberle, P., Eds.; Deutsche Forschungsanstalt fur Lebensmittelchemie: Garching, Germany, 2005; Volume 4953, pp. 213–219. [Google Scholar]
- Sulyok, M.; Beed, F.; Boni, S.; Abass, A.; Mukunzi, A.; Krska, R. Quantitation of multiple mycotoxins and cyanogenic glucosides in cassava samples from Tanzania and Rwanda by an LC-MS/MS-based multi-toxin method. Food Addit. Contam. Part A 2015, 32, 488–502. [Google Scholar] [CrossRef]
- Bashyal, B.P.; Wijeratne, E.M.K.; Faeth, S.H.; Gunatilaka, A.A.L. Globosumones A−C, Cytotoxic Orsellinic Acid Esters from the Sonoran Desert Endophytic Fungus Chaetomium globosum. J. Nat. Prod. 2005, 68, 724–728. [Google Scholar] [CrossRef]
- Schlörke, O.; Zeeck, A. Orsellides A–E: An Example for 6-Deoxyhexose Derivatives Produced by Fungi. Eur. J. Org. Chem. 2006, 2006, 1043–1049. [Google Scholar] [CrossRef]
- Xu, G.-B.; Wang, N.-N.; Bao, J.-K.; Yang, T.; Li, G.-Y. New Orsellinic Acid Esters from Fungus Chaetomium globosporum. Helvetica Chim. Acta 2014, 97, 151–159. [Google Scholar] [CrossRef]
- Nakano, T.; Sugimoto, S.; Matsunami, K.; Otsuka, H. Dianthosaponins A—F, Triterpene Saponins, Flavonoid Glycoside, Aromatic Amide Glucoside and γ-Pyrone Glucoside from Dianthus japonicus. Chem. Pharm. Bull. 2011, 59, 1141–1148. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Y.; Guo, M. Triterpenoid Saponins from the Seeds of Celosia argentea and Their Anti-inflammatory and Antitumor Activities. Chem. Pharm. Bull. 2011, 59, 666–671. [Google Scholar] [CrossRef]
- Sommart, U.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Sakayaroj, J.; Kirtikara, K. Hydronaphthalenones and a Dihydroramulosin from the Endophytic Fungus PSU-N24. Chem. Pharm. Bull. 2008, 56, 1687–1690. [Google Scholar] [CrossRef]
- Gunatilaka, A.A.L. Natural Products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 2006, 69, 509–526. [Google Scholar] [CrossRef]
- Abdalla, M.A.; Matasyoh, J.C. Endophytes as Producers of Peptides: An Overview About the Recently Discovered Peptides from Endophytic Microbes. Nat. Prod. Bioprospecting 2014, 4, 257–270. [Google Scholar] [CrossRef]
- Nisa, H.; Kamili, A.N.; Nawchoo, I.A.; Shafi, S.; Shameem, N.; Bandh, S.A. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015, 82, 50–59. [Google Scholar] [CrossRef]
- Salas, C.E.; Badillo-Corona, J.A.; Ramírez-Sotelo, G.; Oliver-Salvador, C.; Ramí Rez-Sotelo, G. Biologically Active and Antimicrobial Peptides from Plants. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Bondaryk, M.; Staniszewska, M.; Zielińska, P.; Urbańczyk-Lipkowska, Z. Natural Antimicrobial Peptides as Inspiration for Design of a New Generation Antifungal Compounds. J. Fungi 2017, 3, 46. [Google Scholar] [CrossRef]
- Kombrink, A.; Tayyrov, A.; Essig, A.; Stöckli, M.; Micheller, S.; Hintze, J.; van Heuvel, Y.; Dürig, N.; Lin, C.-W.; Kallio, P.T.; et al. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019, 13, 588–602. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Kwak, M.-K.; Liu, R.; Kim, M.-K.; Moon, D.; Kim, A.H.; Song, S.-H.; Kang, S.-O. Cyclic dipeptides from lactic acid bacteria inhibit the proliferation of pathogenic fungi. J. Microbiol. 2014, 52, 64–70. [Google Scholar] [CrossRef]
- Xu, H.; Andi, B.; Qian, J.; West, A.H.; Cook, P.F. The α-Aminoadipate Pathway for Lysine Biosynthesis in Fungi. Cell Biophys. 2006, 46, 43–64. [Google Scholar] [CrossRef]
- Sarjala, T.; Tienaho, J.; Leon-D, E.; Wähälä, K. Antioxidant activity and bioactive properties of root-colonizing endophytic fungi. Coniochaeta lignicola 2019. Manuscript in preparation. [Google Scholar]
- Scott, J.F.; Zamecnik, P.C. SOME OPTICAL PROPERTIES OF DIADENOSINE-5’-PHOSPHATES. Proc. Natl. Acad. Sci. USA 1969, 64, 1308–1314. [Google Scholar] [CrossRef]
- Lucas, K.A.; Filley, J.R.; Erb, J.M.; Graybill, E.R.; Hawes, J.W. Peroxisomal Metabolism of Propionic Acid and Isobutyric Acid in Plants. J. Boil. Chem. 2007, 282, 24980–24989. [Google Scholar] [CrossRef]
- Modess, O. Zur Kenntnis der Mykorrhizabildner von Kiefer und Fichte. In Symbolae botanicae Upsalienses; Anonymous, Ed.; Uppsala Universitet: Uppsala, Sweden, 1941; Volume 5, pp. 1–147. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS. In PCR Protocols; Elsevier BV: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Koukol, O.; Kolařík, M.; Kolářová, Z.; Baldrian, P. Diversity of foliar endophytes in wind-fallen Picea abies trees. Fungal Divers. 2012, 54, 69–77. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


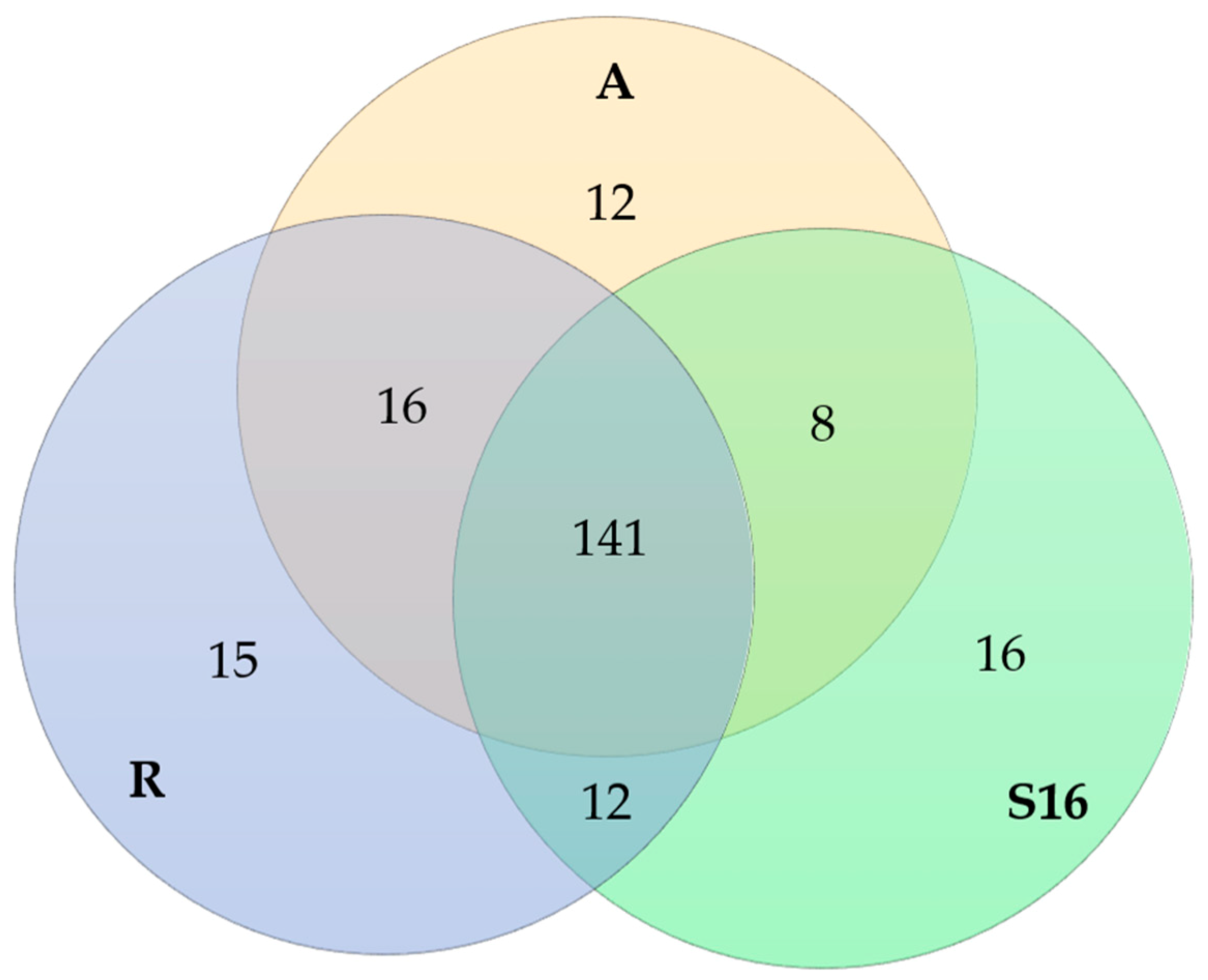
| Strain (GenBank Accession NO.) | GenBank Accession NO. for the Best Match | Max Identity (%)/Query Coverage (%) | Our Description for the Strain | Order | Class | Phylum |
|---|---|---|---|---|---|---|
| A (KM068384) | AY078147.1 | 100/98 | Acephala applanata | Helotiales | Leotiomycetes | Ascomycota |
| R (KJ649992) | AB671499.2 | 100/100 | Phialocephala fortinii | Helotiales | Leotiomycetes | Ascomycota |
| S16 (KJ649998) | KC128659 | 99/98 | Humicolopsis cephalosporioides | Ascomycota | ||
| DQ93680 | 99/98 | Coniochaeta mutabilis | Coniochaetales | Sordariomycetes | Ascomycota |
| Exact Mass | Error | Fungal Extract | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #ID | RT | [M + H]+ | [M − H]− | Measured | Calculated | Δm (ppm) | Molecular Formula | RDB | Compound | Class | MSI | References | A | R | S16 |
| 1 | 0.46 | 175 | 173 | 174.11199 | 174.11168 | 1.8 | C6H14O2N4 | 2 | Arginine | Amino acid | 1 | [23,24,25,26] | x | x | x |
| 2 | 0.49 | 337 | 335 | 336.16316 | 336.16450 | −4.0 | C12H24O7N4 | 3 | Hexosearginine | Amadori | 1 | [27,28,29] | x | x | x |
| 3 | 0.51 | 335 | 333 | 334.06709 | 334.06650 | 1.8 | C9H19O11P | 2 | Glycerophospho-inositol | Common metabolite | 2 | [30,31] | x | - | x |
| 4 | 0.52 | 372 | 370 | 371.22764 | 371.22810 | −1.2 | C15H29O4N7 | 5 | Ace-Ala-Arg-Ala-NMe | Peptide | 2 | x | x | x | |
| 5 | 0.53 | 183 | 181 | 182.07875 | 182.07904 | −1.6 | C6H14O6 | 1 | Mannitol | Hexitol | 1 | [24,26,32] | x | x | x |
| 6 | 0.53 | 246 | 244 | 245.14851 | 245.14879 | −1.1 | C9H19O3N5 | 3 | Ala-Arg or Arg-Ala | Dipeptide | 2 | - | x | x | |
| 7 | 0.54 | nd | 165 | 166.04668 | 166.04774 | −6.4 | C5H10O6 | 1 | Pentonic acid | Pentonic acid | 3 | [26] | x | x | x |
| 8 | 0.54 | nd | 195 | 196.05748 | 196.05831 | −4.2 | C6H12O7 | 1 | Hexonic acid | Hexonic acid | 3 | [26] | x | x | x |
| 9 | 0.54 | 218 | 216 | 217.10586 | 217.10626 | −1.8 | C8H15O4N3 | 3 | Ala-Gln | Dipeptide | 1 | - | - | x | |
| 10 | 0.55 | 203 | 201 | 202.14253 | 202.14298 | −2.2 | C8H18O2N4 | 2 | Dimethylarginine | Amino acid derivative | 2 | [25,33] | x | x | x |
| 11 | 0.55 | 219 | 217 | 218.08984 | 218.09027 | −2.0 | C8H14O5N2 | 3 | Ala-Glu or Glu-Ala or Heliopine | Dipeptide or Opine amino acid | 2 | [34] | - | - | x |
| 12 | 0.55 | 244 | 242 | 243.08558 | 243.08552 | 0.2 | C9H13O5N3 | 5 | Cytidine | Nucleoside | 1 | [25,26,35,36,37,38,39,40] | x | x | x |
| 13 | 0.55 | 248 | 246 | 247.11641 | 247.11682 | −1.7 | C9H17O5N3 | 3 | Gln-Thr or Thr-Gln | Dipeptide | 2 | - | - | x | |
| 14 | 0.55 | 258 | nd | 257.10240 | 257.10282 | −1.6 | C8H20O6NP | 1 | Glycerophosphoryl-choline | Choline | 1 | [25,41] | x | x | x |
| 15 | 0.56 | 191 | 189 | 190.09479 | 190.09536 | −3.0 | C7H14O4N2 | 2 | Ala-Thr or Thr-Ala | Dipeptide | 2 | x | x | x | |
| 16 | 0.56 | 235° | nd | 234.15747 | 234.15796 | −2.1 | C10H22O4N2 | 1 | Valine | Amino acid | 2 | [23,24,26] | - | x | x |
| 17 | 0.56 | 253 | 251 | 252.12182 | 252.12224 | −1.7 | C11H16O3N4 | 6 | His-Pro or Pro-His | Dipeptide | 2 | x | x | x | |
| 18 | 0.56 | 266 | 264 | 265.11662 | 265.11615 | 1.8 | C10H19O7N | 2 | Hexoseaminobutyric acid | Amadori | 3 | [42] | x | x | - |
| 19 | 0.57 | 205 | 203 | 204.11052 | 204.11101 | −2.4 | C8H16O4N2 | 2 | Ser-Val or Val-Ser | Dipeptide | 2 | - | x | x | |
| 20 | 0.58 | 187 | 185 | 186.10007 | 186.10044 | −2.0 | C8H14O3N2 | 3 | Pro-Ala | Dipeptide | 2 | - | - | x | |
| 21 | 0.58 | 198 | 196 | 197.07971 | 197.08004 | −1.7 | C8H11O3N3 | 5 | Acetylhistidine | Amino acid derivative | 2 | - | x | x | |
| 22 | 0.58 | 219 | 217 | 218.12640 | 218.12666 | −1.2 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 3/2 | [34,43] | x | x | x |
| 23 | 0.58 | 246 | 244 | 245.13727 | 245.13756 | −1.2 | C10H19O4N3 | 3 | Dipeptideb | Dipeptide | 3 | x | x | x | |
| 24 | 0.58 | 260 | 258 | 259.18913 | 259.18959 | −1.8 | C12H25O3N3 | 2 | Dipeptidec | Dipeptide | 3 | x | x | x | |
| 25 | 0.59 | 184 | nd | 183.05615 | 183.05653 | −2.1 | C5H13O4NS | 2 | Choline-O-sulphate | Choline | 1 | [41,44] | x | x | x |
| 26 | 0.60 | 233 | 231 | 232.10553 | 232.10592 | −1.7 | C9H16O5N2 | 3 | Asp-Val or Val-Asp | Dipeptide | 2 | x | x | x | |
| 27 | 0.60 | 335 | 333 | 334.06584 | 334.06650 | −2.0 | C9H19O11P | 2 | Glycerophospho-inositol | Common metabolite | 2 | [30,31] | x | x | x |
| 28 | 0.61 | 165 | 163 | 164.06831 | 164.06848 | −1.0 | C6H12O5 | 1 | Fucose | Hexose | 1 | [45] | - | x | - |
| 29 | 0.64 | 191 | 189 | 190.09482 | 190.09536 | −2.8 | C7H14O4N2 | 2 | Ala-Thr or Thr-Ala | Dipeptide | 2 | x | x | - | |
| 30 | 0.64 | 219 | 217 | 218.12619 | 218.12666 | −2.2 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 3/2 | [34,43] | - | x | x |
| 31 | 0.64 | 219 | 217 | 218.08980 | 218.09027 | −2.2 | C8H14O5N2 | 3 | Ala-Glu or Glu-Ala or Heliopine | Dipeptide or Opine amino acid | 2 | [34] | - | - | x |
| 32 | 0.64 | nd | 565 | 566.05592 | 566.05502 | 1.6 | C15H24O17N2P2 | 8 | UDP-galactose | Nucleotide sugar | 2 | [46,47] | x | x | x |
| 33 | 0.65 | 217 | 215 | 216.12187 | 216.12224 | −1.7 | C8H16O3N4 | 3 | Acetylarginine | Amino acid derivative | 2 | [26] | x | x | x |
| 34 | 0.66 | 255 | nd | 254.08980 | 254.09027 | −1.8 | C11H14O5N2 | 6 | Nicotinamide riboside | Pyridine nucleoside | 2 | [48] | x | x | - |
| 35 | 0.67 | 152 | 150 | 151.04916 | 151.04941 | −1.7 | C5H5ON5 | 6 | Guanine | Nucleobase | 1 | [35,38] | - | x | - |
| 36 | 0.67 | 247 | 245 | 246.12114 | 246.12157 | −1.7 | C10H18O5N2 | 3 | Dipeptided | Dipeptide | 3 | x | x | x | |
| 37 | 0.68 | 219 | 217 | 218.08995 | 218.09027 | −1.5 | C8H14O5N2 | 3 | Ala-Glu or Glu-Ala or Heliopine | Dipeptide or Opine amino acid | 2 | [34] | - | - | x |
| 38 | 0.68 | 608 | 606 | 607.08235 | 607.08157 | 1.3 | C17H28O17N3P2 | 8 | UDP-galactosamine | Nucleotide sugar | 2 | [45,49] | x | x | x |
| 39 | 0.69 | 247 | 245 | 246.12112 | 246.12157 | −1.8 | C10H18O5N2 | 3 | Dipeptided | Dipeptide | 3 | x | x | x | |
| 40 | 0.69 | 288 | 286 | 287.19514 | 287.19574 | −2.1 | C12H25O3N5 | 3 | Dipeptidee | Dipeptide | 3 | x | x | x | |
| 41 | 0.71 | 189 | 187 | 188.07948 | 188.07971 | −1.2 | C7H12O4N2 | 3 | Aceglutamide | Amino acid derivative | 2 | x | x | x | |
| 42 | 0.72 | 193 | 191 | 192.02628 | 192.02701 | −3.8 | C6H8O7 | 4 | Isocitrate | Common metabolite | 2 | [50] | x | x | x |
| 43 | 0.72 | 280 | 278 | 279.13127 | 279.1318 | −1.9 | C11H21O7N | 2 | Hexosevaline | Amadori | 2 | [28,29] | - | - | x |
| 44 | 0.73 | 246 | 244 | 245.13705 | 245.13756 | −2.1 | C10H19O4N3 | 3 | Dipeptideb | Dipeptide | 3 | x | x | x | |
| 45 | 0.74 | 348 | 346 | 347.06239 | 347.06309 | −2.0 | C10H14O7N5P | 8 | AMP or dGMP | Nucleotide | 2 | [51,52,53] | - | x | x |
| 46 | 0.74 | nd | 565 | 566.05580 | 566.05502 | 1.4 | C15H24O17N2P2 | 8 | UDP-glucose | Nucleotide sugar | 2 | [46,47,53,54] | x | x | x |
| 47 | 0.77 | 204 | nd | 203.11554 | 203.11576 | −1.1 | C9H17O4N | 2 | Acetylcarnitine | Common metabolite | 2 | [50,55] | x | x | x |
| 48 | 0.77 | 219 | 217 | 218.12622 | 218.12666 | −2.0 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 3/2 | [34,43] | x | x | x |
| 49 | 0.77 | 333 | 331 | 332.13261 | 332.13320 | −1.8 | C12H2O7N4 | 5 | Tripeptide/peptidef | Tripeptide or peptide | 3 | x | x | - | |
| 50 | 0.78 | 205 | 203 | 204.11084 | 204.11101 | −0.8 | C8H16O4N2 | 2 | Ser-Val or Val-Ser | Dipeptide | 2 | x | x | x | |
| 51 | 0.78 | 608 | 606 | 607.08248 | 607.08157 | 1.5 | C17H27O17N3P2 | 8 | UDP-glucoseamine | Nucleotide sugar | 2 | [46,49] | x | x | x |
| 52 | 0.80 | 187 | 185 | 186.10017 | 186.10044 | −1.5 | C8H14O3N2 | 3 | Ala-Pro | Dipeptide | 1 | [26] | x | x | x |
| 53 | 0.80 | 193 | 191 | 192.02628 | 192.02701 | −3.8 | C6H8O7 | 4 | Citric acid | Common metabolite | 1 | [24,26] | x | x | x |
| 54 | 0.80 | 260 | 258 | 259.18922 | 259.18959 | −1.4 | C12H26O3N3 | 2 | Dipeptidec | Dipeptide | 3 | x | x | x | |
| 55 | 0.80 | 315 | 313 | 314.12205 | 314.1213 | 2.4 | C11H22O10 | 1 | Disaccharide | Disaccharide | 3 | [56] | - | x | - |
| 56 | 0.81 | 189 | 187 | 188.11563 | 188.11609 | −2.4 | C8H16O3N2 | 2 | Dipeptideg | Dipeptide | 3 | x | x | x | |
| 57 | 0.81 | 288 | 286 | 287.19523 | 287.19574 | −1.8 | C12H25O3N5 | 3 | Dipeptidee | Dipeptide | 3 | - | x | x | |
| 58 | 0.81 | 333 | 331 | 332.13261 | 332.13320 | −1.8 | C12H20O7N4 | 5 | Tripeptide/peptidef | Tripeptide or peptide | 3 | x | x | - | |
| 59 | 0.82 | 219 | 217 | 218.12630 | 218.12666 | −1.7 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 3/2 | [34,43] | x | - | x |
| 60 | 0.84 | 260 | 258 | 259.15272 | 259.15321 | −1.9 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | x | x | |
| 61 | 0.85 | 219 | 217 | 218.12633 | 218.12666 | −1.5 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 3 | [34,43] | x | x | x |
| 62 | 0.85 | 233 | 231 | 232.14192 | 232.14231 | −1.7 | C10H20O4N2 | 2 | Dipeptidei | Dipeptide | 3 | x | x | x | |
| 63 | 0.85 | 260 | 258 | 259.18870 | 259.18959 | −3.4 | C12H26O3N3 | 2 | Dipeptidec | Dipeptide | 3 | x | x | x | |
| 64 | 0.86 | 221 | 219 | 220.08778 | 220.08817 | −1.8 | C8H16O3N2S | 2 | Cys-Val or Val-Cys or Met-Ala | Dipeptide | 2 | x | x | x | |
| 65 | 0.86 | 245 | 243 | 244.06948 | 244.06954 | −0.2 | C9H12O6N2 | 5 | Pseudouridine | Nucleoside | 2 | [25,35] | - | - | x |
| 66 | 0.86 | 247 | 245 | 246.12114 | 246.12157 | −1.7 | C10H18O5N2 | 3 | Dipeptided | Dipeptide | 3 | x | x | x | |
| 67 | 0.87 | 182 | 180 | 181.07374 | 181.07389 | −0.8 | C9H11O3N | 5 | Tyrosine | Amino acid | 1 | [23,24,25,26,55] | x | x | x |
| 68 | 0.87 | nd | 205 | 206.04209 | 206.04266 | −2.8 | C7H10O7 | 3 | Methylisocitric acid | Common metabolite | 2 | [57,58] | x | x | x |
| 69 | 0.89 | 260 | 258 | 259.15288 | 259.15321 | −1.3 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | x | - | |
| 70 | 0.91 | 190 | 188 | 189.06334 | 189.06372 | −2.0 | C7H11O5N | 3 | Acetylglutamic acid | Amino acid derivative | 2 | - | x | x | |
| 71 | 0.91 | 233 | 231 | 232.14152 | 232.14231 | −3.4 | C10H20O4N2 | 2 | Dipeptidei | Dipeptide | 3 | x | x | x | |
| 72 | 0.91 | 260 | 258 | 259.18907 | 259.18959 | −2.0 | C12H26O3N3 | 2 | Dipeptidec | Dipeptide | 3 | x | x | x | |
| 73 | 0.93 | 233 | 231 | 232.10574 | 232.10592 | −0.8 | C9H16O5N2 | 3 | Asp-Val or Val-Asp | Dipeptide | 2 | x | x | x | |
| 74 | 0.95 | nd | 205 | 206.04215 | 206.04266 | −2.5 | C7H10O7 | 4 | Methylcitric acid | Common metabolite | 2 | [57,58] | x | x | x |
| 75 | 0.95 | 218 | 216 | 217.10599 | 217.10626 | −1.2 | C8H15O4N3 | 3 | Acetylcitrulline | Amino acid derivative | 2 | - | x | - | |
| 76 | 0.97 | 190 | 188 | 189.06359 | 189.06372 | −0.7 | C7H11O5N | 3 | Acetylglutamic acid | Amino acid derivative | 2 | - | - | x | |
| 77 | 0.97 | 307 | 305 | 306.02467 | 306.02530 | −2.1 | C9H11O8N2P | 7 | 2’,3’-cUMP or 3’,5’-cUMP | Nucleotide | 2 | [59,60,61,62] | - | - | x |
| 78 | 0.99 | 245 | 243 | 244.06915 | 244.06954 | −1.6 | C9H12O6N2 | 5 | Uridine | Nucleoside | 1 | [35,36,38,39,63] | x | x | x |
| 79 | 0.99 | 288 | 286 | 287.19481 | 287.19574 | −3.2 | C12H25O3N5 | 3 | Dipeptidee | Dipeptide | 3 | x | x | x | |
| 80 | 0.99 | 332 | 330 | 331.06749 | 331.06817 | −2.0 | C10H14O6N5P | 8 | dAMP | Nucleotide | 2 | [52,53,64] | - | x | x |
| 81 | 1.01 | 253 | 251 | 252.11056 | 252.11101 | −1.8 | C12H16O4N2 | 6 | Ala-Tyr | Dipeptide | 1 | x | x | x | |
| 82 | 1.03 | 261 | 259 | 260.13679 | 260.13722 | −1.6 | C11H20O5N2 | 3 | Dipeptidej | Dipeptide | 3 | x | x | x | |
| 83 | 1.04 | 203 | 201 | 202.13162 | 202.13174 | −0.6 | C9H18O3N2 | 2 | Leu-Ala | Dipeptide | 1 | x | x | x | |
| 84 | 1.04 | 215 | 213 | 214.13141 | 214.13174 | −1.5 | C10H18O3N2 | 3 | Pro-Val or Val-Pro | Dipeptide | 2 | x | x | x | |
| 85 | 1.08 | 221 | 219 | 220.08783 | 220.08817 | −1.5 | C8H16O3N2S | 2 | Cys-Val or Val-Cys or Met-Ala | Dipeptide | 2 | x | x | x | |
| 86 | 1.10 | 261 | 259 | 260.13679 | 260.13722 | −1.6 | C11H20O5N2 | 3 | Dipeptidej | Dipeptide | 3 | x | x | x | |
| 87 | 1.10 | 664 | 662 | 663.10777 | 663.10912 | −2.0 | C21H27O14N7P2 | 15 | Nicotinamide adenine dinucleotide | Dinucleotide | 2 | [48] | - | - | x |
| 88 | 1.11 | 260 | 258 | 259.15306 | 259.15321 | −0.6 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | - | - | |
| 89 | 1.17 | 202 | nd | 201.11101 | 201.11134 | −1.6 | C8H15O3N3 | 3 | Acetylmethyl-glutaminamide | Amino acid derivative | 2 | x | - | - | |
| 90 | 1.17 | 403 | 401 | 402.22194 | 402.22268 | −1.8 | C16H30O6N6 | 5 | Tripeptide/peptidek | Tripeptide or peptide | 3 | x | - | - | |
| 91 | 1.18 | 613 | 611 | 612.15080 | 612.15197 | −1.9 | C20H32O12N6S2 | 8 | Glutathione dimer | Tripeptide | 2 | [17,55,65] | x | - | x |
| 92 | 1.23 | 315 | 313 | 314.12193 | 314.12130 | 2.0 | C11H22O10 | 1 | Disaccharide | Disaccharide | 3 | [56] | x | x | - |
| 93 | 1.24 | 189 | 187 | 188.11571 | 188.11609 | −2.0 | C8H16O3N2 | 2 | Dipeptideg | Dipeptide | 3 | x | x | x | |
| 94 | 1.24 | 269 | 267 | 268.10536 | 268.10592 | −2.1 | C12H16O5N2 | 6 | Ser-Tyr or Tyr-Ser | Dipeptides | 2 | x | x | x | |
| 95 | 1.30 | 268 | 266 | 267.09608 | 267.09676 | −2.5 | C10H13O4N5 | 7 | Adenosine | Nucleoside | 1 | [25,26,35,36,37,38,40,55,63] | x | x | x |
| 96 | 1.33 | 315 | 313 | 314.12166 | 314.12130 | 1.2 | C11H22O10 | 1 | Disaccharide | Disaccharide | 3 | [56] | - | x | - |
| 97 | 1.33 | 542 | 540 | 541.05998 | 541.06111 | −2.1 | C15H21O13N5P2 | 11 | Cyclic ADP-ribose | Nucleotide | 2 | [66] | x | x | - |
| 98 | 1.40 | 179 | 177 | 178.04742 | 178.04774 | −1.8 | C6H10O6 | 2 | Dehydrohexose | Dehydro-hexose | 3 | [67] | x | x | x |
| 99 | 1.42 | 221 | 219 | 220.08820 | 220.08817 | 0.1 | C8H16O3N2S | 2 | Ala-Met | Dipeptide | 1 | - | - | x | |
| 100 | 1.47 | 346 | 344 | 345.04695 | 345.04744 | −1.4 | C10H12O7N5P | 9 | 2’,3’-cGMP or 3’,5’-cGMP | Nucleotide | 2 | [25,38,51,59,60,61,62] | x | x | x |
| 101 | 1.48 | 219 | 217 | 218.12633 | 218.12666 | −1.5 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 2/3 | [34,43] | x | x | x |
| 102 | 1.50 | 267 | 265 | 266.12611 | 266.12666 | −2.1 | C13H18O4N2 | 6 | Dipeptidel | Dipeptide | 3 | x | x | x | |
| 103 | 1.50 | 284 | 282 | 283.09056 | 283.09167 | −3.9 | C10H13O5N5 | 7 | Guanosine isomer | Nucleoside | 2 | [26,35,37,39] | x | x | x |
| 104 | 1.54 | 346 | 344 | 345.04701 | 345.04744 | −1.2 | C10H12O7N5P | 9 | 2’,3’-cGMP or 3’,5’-cGMP | Nucleotide | 2 | [25,38,51,59,60,61,62] | x | - | x |
| 105 | 1.57 | 165 | 163 | 164.06821 | 164.06848 | −1.6 | C6H12O5 | 1 | Deoxyhexose | Deoxyhexose | 3 | [68] | x | x | - |
| 106 | 1.61 | 246 | 244 | 245.13713 | 245.13756 | −1.7 | C10H19O4N3 | 3 | Dipeptideb | Dipeptide | 3 | x | x | x | |
| 107 | 1.64 | 219 | 217 | 218.12628 | 218.12666 | −1.7 | C9H18O4N2 | 2 | Dipeptidea or Lysopine or Rideopine | Dipeptide or Opine amino acid | 3/2 | [34,43] | x | x | x |
| 108 | 1.64 | 267 | 265 | 266.12620 | 266.12666 | −1.7 | C13H18O4N2 | 6 | Dipeptidel | Dipeptide | 3 | x | x | x | |
| 109 | 1.67 | 260 | 258 | 259.15294 | 259.15321 | −1.0 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | x | x | |
| 110 | 1.68 | 294 | 292 | 293.13692 | 293.13756 | −2.2 | C14H19O4N3 | 7 | Di/tripeptidem | Di-/Tripeptide | 3 | x | x | x | |
| 111 | 1.69 | 233 | 231 | 232.14200 | 232.14231 | −1.3 | C10H20O4N2 | 2 | Dipeptidei | Dipeptide | 3 | x | x | x | |
| 112 | 1.71 | 281 | 279 | 280.10530 | 280.10592 | −2.2 | C13H16O5N2 | 7 | Abenquine C or enantiomer | AA quinone | 2 | [69,70] | x | x | x |
| 113 | 1.72 | 189 | 187 | 188.11572 | 188.11609 | −2.0 | C8H16O3N2 | 2 | Dipeptideg | Dipeptide | 3 | x | x | x | |
| 114 | 1.74 | 152 | 150 | 151.04925 | 151.04941 | −1.0 | C5H5ON5 | 6 | Isoguanine or Oxyadenine | Nucleobase | 2 | [71,72] | - | x | x |
| 115 | 1.74 | 246 | 244 | 245.13727 | 245.13756 | −1.2 | C10H19O4N3 | 3 | Dipeptideb | Dipeptide | 3 | x | x | x | |
| 116 | 1.74 | 279 | 277 | 278.12642 | 278.12666 | −0.9 | C14H18O4N2 | 7 | Pro-Tyr or Tyr-Pro | Dipeptide | 2 | x | x | x | |
| 117 | 1.74 | 288 | 286 | 287.19527 | 287.19574 | −1.6 | C12H25O3N5 | 3 | Dipeptidee | Dipeptide | 3 | - | - | x | |
| 118 | 1.75 | 261 | 259 | 260.13698 | 260.13722 | −0.9 | C11H20O5N2 | 3 | Fusarinine monomer | Siderophore | 2 | [73,74,75,76,77] | x | x | x |
| 119 | 1.75 | 268 | 266 | 267.09639 | 267.09676 | −1.4 | C10H13O4N5 | 7 | Deoxyguanosine | Nucleoside | 2 | [35] | x | x | x |
| 120 | 1.75 | 318 | 316 | 317.15840 | 317.15869 | −0.9 | C13H23O6N3 | 4 | Tripeptide/peptiden | Tripeptide/ peptide | 3 | x | x | x | |
| 121 | 1.76 | 166 | 164 | 165.07893 | 165.07898 | −0.3 | C9H11O2N | 5 | Phenylalanine | Amino acid | 1 | [23,24,26,55] | x | x | x |
| 122 | 1.76 | 189 | 187 | 188.11574 | 188.11609 | −1.8 | C8H16O3N2 | 2 | Dipeptideg | Dipeptide | 3 | - | - | x | |
| 123 | 1.76 | 260 | 258 | 259.15306 | 259.15321 | −0.6 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | x | x | |
| 124 | 1.78 | 257 | 255 | 256.10554 | 256.10592 | −1.5 | C11H16O5N2 | 5 | Methylthymidine | Nucleoside derivative | 2 | [78] | x | x | x |
| 125 | 1.78 | 266 | 264 | 265.11577 | 265.11615 | −1.4 | C10H19O7N | 2 | Deoxyhexose-threonine | Amadori | 3 | [79] | x | x | - |
| 126 | 1.80 | 203 | 201 | 202.13164 | 202.13174 | −0.5 | C9H18O3N2 | 2 | Ala-Leu | Dipeptide | 1 | x | x | x | |
| 127 | 1.80 | 260 | 258 | 259.15303 | 259.15321 | −0.7 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | x | x | |
| 128 | 1.82 | 189 | 187 | 188.11586 | 188.11609 | −1.2 | C8H16O3N2 | 2 | Dipeptideg | Dipeptide | 3 | x | x | x | |
| 129 | 1.82 | 215 | 213 | 214.13157 | 214.13174 | −0.8 | C10H18O3N2 | 3 | Pro-Val or Val-Pro | Dipeptide | 2 | x | x | x | |
| 130 | 1.82 | 233 | 231 | 232.14207 | 232.14231 | −1.0 | C10H20O4N2 | 2 | Dipeptidei | Dipeptide | 3 | x | x | x | |
| 131 | 1.85 | 307 | 305 | 306.02538 | 306.02530 | 0.3 | C9H11O8N2P | 7 | 2’,3’-cUMP or 3’,5’-cUMP | Nucleotide | 2 | [59,60,61,62] | x | x | x |
| 132 | 1.86 | 260 | 258 | 259.15312 | 259.15321 | −0.3 | C11H21O4N3 | 3 | Di-/Tripeptideh | Di-/Tripeptide | 3 | x | - | x | |
| 133 | 1.86 | 348 | 346 | 347.16926 | 347.16925 | 0.0 | C14H25O7N3 | 4 | Tripeptideo | Tripeptide | 3 | x | - | - | |
| 134 | 1.87 | 249 | 247 | 248.11934 | 248.11947 | −0.5 | C10H20O3N2S | 2 | Met-Val or Val-Met | Dipeptide | 2 | x | x | x | |
| 135 | 1.89 | 261 | 259 | 260.13682 | 260.13722 | −1.5 | C11H20O5N2 | 3 | Fusarinine monomer | Siderophore | 2 | [73,74,75,76,77] | x | x | x |
| 136 | 1.89 | 281 | 279 | 280.14177 | 280.14231 | −1.9 | C14H20O4N2 | 6 | Val-Tyr | Dipeptide | 1 | x | x | x | |
| 137 | 1.89 | 295 | 293 | 294.12105 | 294.12157 | −1.8 | C14H18O5N2 | 7 | Peptide type compoundp | Dipeptide or peptide derivative | 3 | [69,70] | x | x | x |
| 138 | 1.90 | 318 | 316 | 317.15846 | 317.15869 | −0.7 | C13H23O6N3 | 4 | Tripeptide/peptiden | Tripeptide/ peptide | 3 | x | - | - | |
| 139 | 1.90 | 317 | 315 | 316.09129 | 316.09067 | 2.0 | C12H16O8N2 | 7 | 5-methoxycarbonyl-methyluridine | Nucleoside derivative | 2 | [39,40] | x | - | - |
| 140 | 1.91 | 229 | 227 | 228.14731 | 228.14739 | −0.3 | C11H20O3N2 | 3 | Pro-Leu | Dipeptide | 1 | x | x | x | |
| 141 | 1.91 | 247 | 245 | 246.12143 | 246.12157 | −0.6 | C10H18O5N2 | 3 | Dipeptided | Dipeptide | 3 | x | x | x | |
| 142 | 1.95 | 249 | 247 | 248.11934 | 248.11947 | −0.5 | C10H20O3N2S | 2 | Met-Val or Val-Met | Dipeptide | 2 | x | x | x | |
| 143 | 1.96 | 231 | 229 | 230.16318 | 230.16304 | 0.6 | C11H22O3N2 | 2 | Dipeptideq | Dipeptide | 3 | x | x | x | |
| 144 | 1.96 | 237 | 235 | 236.11595 | 236.11609 | −0.6 | C12H16O3N2 | 6 | Ala-Phe | Dipeptide | 1 | x | x | x | |
| 145 | 1.96 | 348 | 346 | 347.16933 | 347.16925 | 0.2 | C14H25O7N3 | 4 | Tripeptideo | Tripeptide | 3 | x | x | - | |
| 146 | 1.99 | 223 | 221 | 222.10043 | 222.10044 | 0.0 | C11H14O3N2 | 6 | Gly-Phe | Dipeptide | 1 | x | x | x | |
| 147 | 1.99 | 246 | 244 | 245.13786 | 245.13756 | 1.2 | C10H19O4N3 | 3 | Dipeptideb | Dipeptide | 3 | x | - | - | |
| 148 | 2.02 | 541 | 539 | 540.14854 | 540.14791 | 1.2 | C24H28O14 | 11 | Phomone A or B or Blumeoside C | Endophyte or plant metabolite | 2 | [80,81,82] | - | x | x |
| 149 | 2.06 | 355 | 353 | 354.15419 | 354.15394 | 0.7 | C15H22O6N4 | 7 | Peptider | Peptide | 3 | - | - | x | |
| 150 | 2.07 | 581 | 579 | 580.15391 | 580.15436 | −0.8 | C20H25O9N10P | 15 | Dinucleotide | Dinucleotide | 3 | - | x | - | |
| 151 | 2.08 | 229 | 227 | 228.14738 | 228.14739 | 0.0 | C11H20O3N2 | 3 | Ile-Pro or Pro-Ile | Dipeptide | 2 | x | x | x | |
| 152 | 2.09 | 231 | 229 | 230.16290 | 230.16304 | −0.6 | C11H22O3N2 | 2 | Dipeptideq | Dipeptide | 3 | x | x | x | |
| 153 | 2.10 | 247 | 245 | 246.12100 | 246.12157 | −2.3 | C10H18O5N2 | 3 | Dipeptided | Dipeptide | 3 | x | x | x | |
| 154 | 2.10 | 306 | 304 | 305.13731 | 305.13756 | −0.8 | C15H19O4N3 | 8 | Thr-Trp or Trp-Thr | Dipeptide | 2 | x | x | x | |
| 155 | 2.10 | 581 | 579 | 580.15428 | 580.15436 | −0.1 | C20H25O9N10P | 15 | Dinucleotide | Dinucleotide | 3 | - | x | - | |
| 156 | 2.14 | 318 | 316 | 317.15843 | 317.15869 | −0.8 | C13H23O6N3 | 4 | Tripeptide/peptiden | Tripeptide/ peptide | 3 | x | x | x | |
| 157 | 2.15 | 253 | 251 | 252.11087 | 252.11101 | −0.5 | C12H16O4N2 | 6 | Phe-Ser or Ser-Phe | Dipeptide | 2 | [55] | x | x | x |
| 158 | 2.15 | 279 | 277 | 278.12630 | 278.12666 | −1.3 | C14H18O4N2 | 7 | Pro-Tyr or Tyr-Pro | Dipeptide | 2 | x | x | x | |
| 159 | 2.19 | 455 | 453 | 454.15532 | 454.15560 | −0.6 | C16H30O7N4S2 | 4 | Peptides | Peptide | 3 | x | x | - | |
| 160 | 2.20 | 280 | 278 | 279.12166 | 279.12191 | −0.9 | C13H17O4N3 | 7 | Di/tripeptidet | Di-/Tripeptide | 3 | x | x | x | |
| 161 | 2.20 | 318 | 316 | 317.15861 | 317.15869 | −0.2 | C13H23O6N3 | 4 | Tripeptide/peptiden | Tripeptide/ peptide | 3 | x | x | x | |
| 162 | 2.22 | 229 | 227 | 228.14723 | 228.14739 | −0.7 | C11H20O3N2 | 3 | Leu-Pro | Dipeptide | 1 | x | x | x | |
| 163 | 2.22 | 345 | 343 | 344.13676 | 344.13722 | −1.3 | C18H20O5N2 | 10 | Tyr-Tyr | Dipeptide | 2 | x | x | x | |
| 164 | 2.22 | 348 | 346 | 347.16899 | 347.16925 | −0.7 | C14H25O7N3 | 4 | Tripeptideo | Tripeptide | 3 | x | - | - | |
| 165 | 2.24 | 223 | 221 | 222.10048 | 222.10044 | 0.2 | C11H14O3N2 | 6 | Phe-Gly | Dipeptide | 2 | x | x | x | |
| 166 | 2.26 | 277 | 275 | 276.11095 | 276.11101 | −0.2 | C14H16O4N2 | 8 | Peptide type compoundu | Dipeptide/ cyclodipeptide | 3 | [83,84] | x | x | x |
| 167 | 2.27 | 294 | 292 | 293.13713 | 293.13756 | −1.5 | C14H19O4N3 | 7 | Di/tripeptidem | Di-/Tripeptide | 3 | x | x | x | |
| 168 | 2.27 | 295 | 293 | 294.15724 | 294.15796 | −2.4 | C15H22O4N2 | 6 | Leu-Tyr | Dipeptide | 1 | x | x | x | |
| 169 | 2.28 | 247 | 245 | 246.12108 | 246.12157 | −2.0 | C10H18O5N2 | 3 | Dipeptided | Dipeptide | 3 | x | x | x | |
| 170 | 2.29 | 231 | 229 | 230.16275 | 230.16304 | −1.2 | C11H22O3N2 | 2 | Dipeptideq | Dipeptide | 3 | x | x | x | |
| 171 | 2.29 | 237 | 235 | 236.11557 | 236.11609 | −2.2 | C12H16O3N2 | 6 | Gly-Phe or Phe-Gly methyl ester | Dipeptide derivative | 2 | x | x | x | |
| 172 | 2.30 | 267 | 265 | 266.12630 | 266.12666 | −1.3 | C13H18O4N2 | 6 | Dipeptidel | Dipeptide | 3 | x | x | x | |
| 173 | 2.32 | 229 | 227 | 228.14734 | 228.14739 | −0.2 | C11H20O3N2 | 3 | Ile-Pro or Pro-Ile | Dipeptide | 2 | x | x | x | |
| 174 | 2.32 | 557 | 555 | 556.15074 | 556.14283 | 14.2 | C24H28O15 | 11 | Blumeoside A | Plant metabolite | 2 | [80] | - | x | - |
| 175 | 2.34 | 205 | 203 | 204.08986 | 204.08988 | −0.1 | C11H12O2N2 | 7 | Tryptophan | Amino acid | 1 | [23,24,25,26,55] | x | x | x |
| 176 | 2.34 | 227 | 225 | 226.09527 | 226.09536 | −0.4 | C10H14O4N2 | 5 | Deoxythymidine | Nucleoside | 2 | [85] | x | - | x |
| 177 | 2.34 | 263 | 261 | 262.13484 | 262.13512 | −1.1 | C11H22O3N2S | 2 | Dipeptidev | Dipeptide | 3 | x | x | x | |
| 178 | 2.35 | 261 | 259 | 260.13704 | 260.13722 | −0.7 | C11H20O5N2 | 3 | Dipeptidej | Dipeptide | 3 | x | x | x | |
| 179 | 2.37 | 295 | 293 | 294.15764 | 294.15796 | −1.1 | C15H22O4N2 | 6 | Dipeptidew | Dipeptide | 3 | x | x | x | |
| 180 | 2.38 | 281 | 279 | 280.10551 | 280.10592 | −1.5 | C13H16O5N2 | 7 | Abenquine C or enantiomer | AA quinone | 2 | [69,70] | x | x | x |
| 181 | 2.39 | 277 | 275 | 276.11095 | 276.11101 | −0.2 | C14H16O4N2 | 8 | Peptide type compoundu | Dipeptide/ cyclodipeptide | 3 | [83,84] | x | x | x |
| 182 | 2.39 | 295 | 293 | 294.12163 | 294.12157 | 0.2 | C14H18O5N2 | 7 | Peptide type compoundp | Dipeptide or peptide derivative | 3 | [69,70] | x | x | x |
| 183 | 2.40 | 160 | 158 | 159.08949 | 159.08954 | −0.3 | C7H13O3N | 2 | Acetylvaline | Amino acid derivative | 2 | [26] | x | x | - |
| 184 | 2.41 | 276 | 274 | 275.12709 | 275.12699 | 0.4 | C14H17O3N3 | 8 | Ala-Trp | Dipeptide | 1 | x | x | x | |
| 185 | 2.45 | 261 | 259 | 260.13728 | 260.13722 | 0.2 | C11H20O5N2 | 3 | Dipeptidej | Dipeptide | 3 | x | x | x | |
| 186 | 2.45 | 263 | 261 | 262.13499 | 262.13512 | −0.5 | C11H22O3N2S | 2 | Dipeptidev | Dipeptide | 3 | x | x | x | |
| 187 | 2.46 | 295 | 293 | 294.15803 | 294.15796 | 0.2 | C15H22O4N2 | 6 | Dipeptidew | Dipeptide | 3 | x | x | x | |
| 188 | 2.50 | 281 | 279 | 280.10603 | 280.10592 | 0.4 | C13H16O5N2 | 7 | α-Asp-Phe | Dipeptide | 1 | x | x | x | |
| 189 | 2.54 | 245 | 243 | 244.17876 | 244.17869 | 0.3 | C12H24O3N2 | 2 | Leu-Leu | Dipeptide | 1 | x | x | x | |
| 190 | 2.54 | 281 | 279 | 280.10582 | 280.10592 | −0.3 | C13H16O5N2 | 7 | β-Asp-Phe | Dipeptide | 1 | x | x | - | |
| 191 | 2.55 | 263 | 261 | 262.13402 | 262.13512 | −4.2 | C11H22O3N2S | 2 | Dipeptidev | Dipeptide | 3 | x | x | x | |
| 192 | 2.55 | 265 | 263 | 264.14735 | 264.14739 | −0.1 | C14H20O3N2 | 6 | Val-Phe or Phe-Val | Dipeptide | 2 | x | x | x | |
| 193 | 2.58 | 253 | 251 | 252.12127 | 252.12224 | −3.8 | C11H16O3N4 | 6 | His-Pro or Pro-His | Dipeptide | 2 | x | x | x | |
| 194 | 2.59 | 382 | 380 | 381.15388 | 381.15360 | 0.7 | C17H23O7N3 | 8 | Tripeptidex | Tripeptide | 3 | x | x | - | |
| 195 | 2.60 | 293 | 291 | 292.10606 | 292.10592 | 0.5 | C14H16O5N2 | 8 | Pyr-Tyr or Cyclo(Glu-Tyr) | Dipeptide/ cyclo-dipeptide | 2 | [83] | x | - | - |
| 196 | 2.61 | 192 | 190 | 191.06101 | 191.06162 | −3.2 | C7H13O3NS | 2 | Acetylmethionine | Amino acid derivative | 2 | [26,86] | - | x | x |
| 197 | 2.66 | 281 | 279 | 280.10557 | 280.10592 | −1.2 | C13H16O5N2 | 7 | Phe-Asp | Dipeptide | 2 | x | x | x | |
| 198 | 2.66 | 295 | 293 | 294.15748 | 294.15796 | −1.6 | C15H22O4N2 | 6 | Dipeptidew | Dipeptide | 3 | x | - | x | |
| 199 | 2.69 | 263 | 261 | 262.13505 | 262.13512 | −0.3 | C11H22O3N2S | 2 | Dipeptidev | Dipeptide | 3 | x | x | x | |
| 200 | 2.75 | 276 | 274 | 275.12715 | 275.12699 | 0.6 | C14H17O3N3 | 8 | Trp-Ala | Dipeptide | 2 | x | x | x | |
| 201 | 2.75 | 433 | 431 | 432.12736 | 432.12678 | 1.3 | C18H24O12 | 7 | Asperulosidic acid or isomer | Plant metabolite | 2 | [87,88,89,90,91] | - | x | - |
| 202 | 2.75 | 518 | 516 | 517.23808 | 517.23840 | −0.6 | C21H35O10N5 | 7 | Peptidey | Peptide | 3 | x | x | - | |
| 203 | 2.77 | 306 | 304 | 305.13740 | 305.13756 | −0.5 | C15H19O4N3 | 8 | Thr-Trp or Trp-Thr | Dipeptide | 2 | x | x | x | |
| 204 | 2.78 | 295 | 293 | 294.12150 | 294.12157 | −0.2 | C14H18O5N2 | 7 | Peptide type compoundp | Dipeptide or peptide derivative | 3 | [69,70] | x | x | x |
| 205 | 2.81 | 265 | 263 | 264.14732 | 264.14739 | −0.3 | C14H20O3N2 | 6 | Val-Phe or Phe-Val | Dipeptide | 2 | x | x | x | |
| 206 | 2.91 | 248 | 246 | 247.10475 | 247.10559 | −3.4 | C10H17O6N | 3 | Pentoseproline or Valinopine or Linamarin | Amadori or Opine amino acid or Plant metabolite | 2 | [92,93,94] | x | x | x |
| 207 | 2.99 | 287 | 285 | 286.10512 | 286.10526 | −0.5 | C13H18O7 | 5 | Orsellinic acid ester | Endophytic fungi metabolite | 3 | [16,95,96,97] | - | x | - |
| 208 | 3.01 | 248 | 246 | 247.10585 | 247.10559 | 1.1 | C10H17O6N | 3 | Pentoseproline or Valinopine or Linamarin | Amadori or Opine amino acid or Plant metabolite | 2 | [92,93,94] | x | x | x |
| 209 | 3.02 | 528 | 526 | 527.29613 | 527.29552 | 1.2 | C24H41O8N5 | 7 | Peptidez | Peptide | 3 | - | x | - | |
| 210 | 3.11 | 243 | 241 | 242.12691 | 242.12666 | 1.0 | C11H18O4N2 | 4 | Dipeptide# | Dipeptide | 3 | x | - | - | |
| 211 | 3.22 | 174 | 172 | 173.10513 | 173.10519 | −0.3 | C8H15O3N | 2 | Acetylleucine or acetylisoleucine | Amino acid derivative | 2 | [26] | - | x | - |
| 212 | 3.29 | 243 | 241 | 242.12608 | 242.12666 | −2.4 | C11H18O4N2 | 4 | Dipeptide# | Dipeptide | 3 | x | - | - | |
| 213 | 3.37 | 174 | 172 | 173.10538 | 173.10519 | 1.1 | C8H15O3N | 2 | Acetylleucine or acetylisoleucine | Amino acid derivative | 2 | [26] | - | x | - |
| 214 | 3.37 | 663 | 661 | 662.33214 | 662.33023 | 2.9 | C35H50O12 | 11 | Saponin | Saponin | 3 | [98,99] | - | - | x |
| 215 | 3.50 | 282 | 280 | 281.08955 | 281.08994 | −1.4 | C13H15O6N | 7 | Phenylacetyl-glutamine | Amino acid derivative | 2 | x | x | x | |
| 216 | 3.76 | 277 | 275 | 276.11052 | 276.11101 | −1.8 | C14H16O4N2 | 8 | Peptide type compoundu | Dipeptide/ cyclodipeptide | 3 | [83,84] | x | - | - |
| 217 | 4.07 | 517 | 515 | 516.27897 | 516.27685 | 4.1 | C19H36O7N10 | 7 | Peptide¤ | Peptide | 3 | - | x | - | |
| 218 | 4.23 | 247 | 245 | 246.10060 | 246.10044 | 0.7 | C13H14O3N2 | 8 | Acetyltryptophan | Amino acid derivative | 2 | x | - | x | |
| 219 | 4.30 | 397 | 395 | 396.21529 | 396.21212 | 8.0 | C17H28O5N6 | 6 | Tripeptide/peptide§ | Tripeptide/ peptide | 3 | - | x | - | |
| 220 | 4.95 | 201 | 199 | 200.10441 | 200.10486 | −2.2 | C10H16O4 | 2 | Ramulosin derivative | endophytic fungi metabolite | 3 | [16,100] | x | x | x |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tienaho, J.; Karonen, M.; Muilu–Mäkelä, R.; Wähälä, K.; Leon Denegri, E.; Franzén, R.; Karp, M.; Santala, V.; Sarjala, T. Metabolic Profiling of Water-Soluble Compounds from the Extracts of Dark Septate Endophytic Fungi (DSE) Isolated from Scots Pine (Pinus sylvestris L.) Seedlings Using UPLC–Orbitrap–MS. Molecules 2019, 24, 2330. https://doi.org/10.3390/molecules24122330
Tienaho J, Karonen M, Muilu–Mäkelä R, Wähälä K, Leon Denegri E, Franzén R, Karp M, Santala V, Sarjala T. Metabolic Profiling of Water-Soluble Compounds from the Extracts of Dark Septate Endophytic Fungi (DSE) Isolated from Scots Pine (Pinus sylvestris L.) Seedlings Using UPLC–Orbitrap–MS. Molecules. 2019; 24(12):2330. https://doi.org/10.3390/molecules24122330
Chicago/Turabian StyleTienaho, Jenni, Maarit Karonen, Riina Muilu–Mäkelä, Kristiina Wähälä, Eduardo Leon Denegri, Robert Franzén, Matti Karp, Ville Santala, and Tytti Sarjala. 2019. "Metabolic Profiling of Water-Soluble Compounds from the Extracts of Dark Septate Endophytic Fungi (DSE) Isolated from Scots Pine (Pinus sylvestris L.) Seedlings Using UPLC–Orbitrap–MS" Molecules 24, no. 12: 2330. https://doi.org/10.3390/molecules24122330
APA StyleTienaho, J., Karonen, M., Muilu–Mäkelä, R., Wähälä, K., Leon Denegri, E., Franzén, R., Karp, M., Santala, V., & Sarjala, T. (2019). Metabolic Profiling of Water-Soluble Compounds from the Extracts of Dark Septate Endophytic Fungi (DSE) Isolated from Scots Pine (Pinus sylvestris L.) Seedlings Using UPLC–Orbitrap–MS. Molecules, 24(12), 2330. https://doi.org/10.3390/molecules24122330