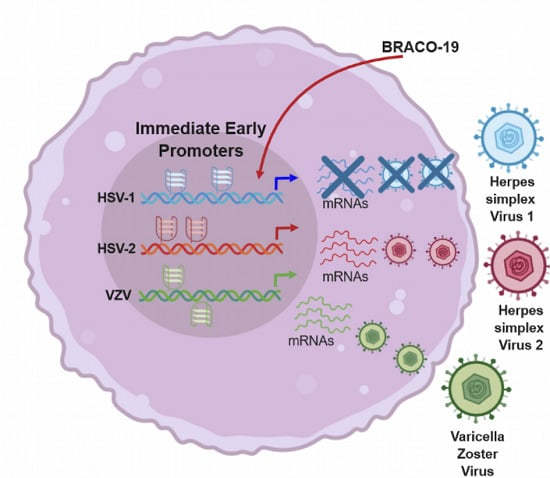
Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses
Abstract
:1. Introduction
2. Results
2.1. Detection of PQS in Herpesviridae Immediate Early (IE) Promoters
2.2. The Identified PQSs Fold into G4 Structures
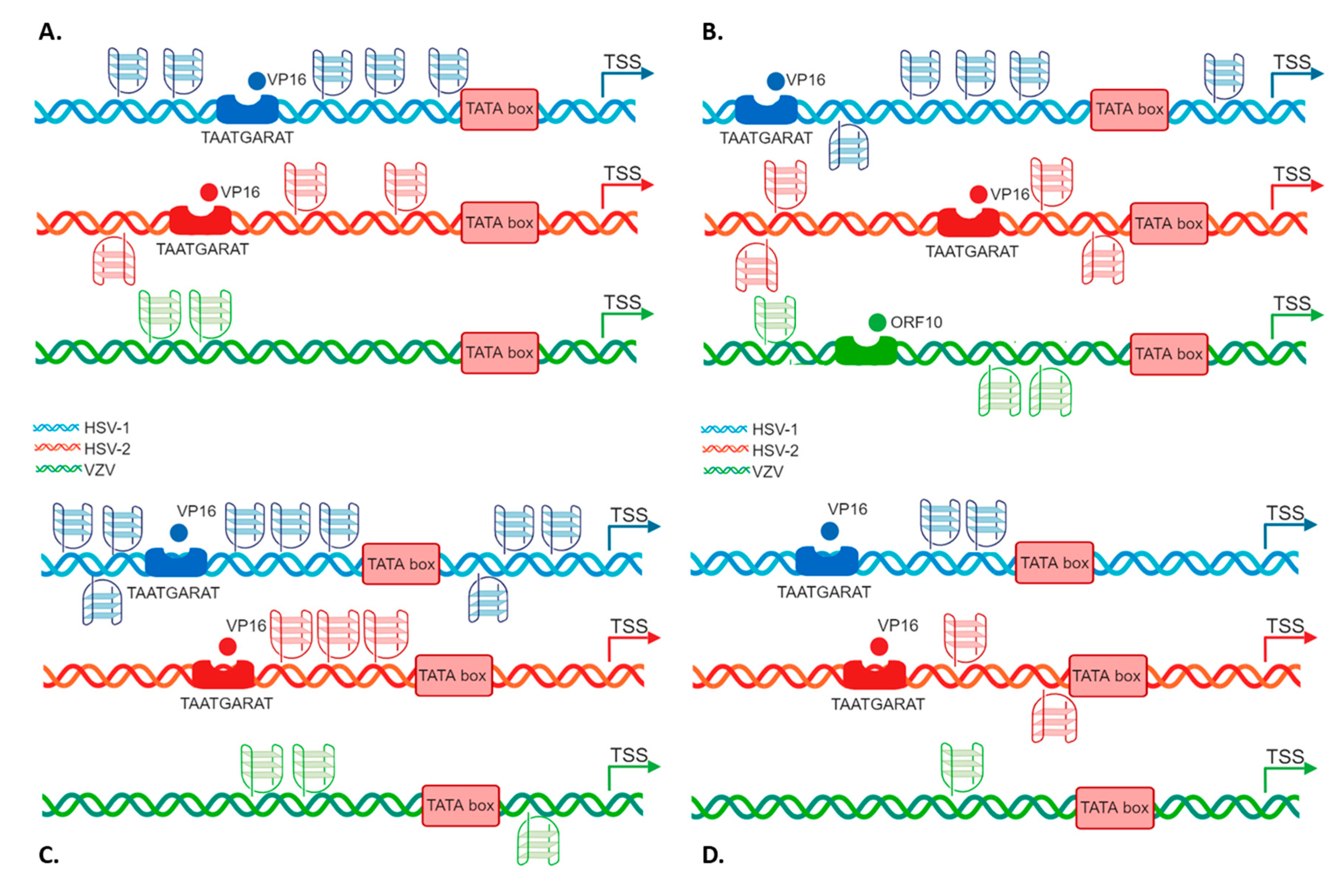
2.3. G4s Tune IE Promoter Activity at the Cellular Level
3. Discussion
4. Materials and Methods
4.1. PQS Detection and Evaluation of Conservation
4.2. Oligonucleotides and Cell Lines
4.3. Circular Dichroism Spectroscopy
4.4. Taq Polymerase Stop Assay
4.5. Plasmids Construction
4.6. Reporter Assays
4.7. Cellular Cytotoxicity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cutter, A.R.; Hayes, J.J. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Bacolla, A.; Wang, G.; Vasquez, K.M. New Perspectives on DNA and RNA Triplexes As Effectors of Biological Activity. PLoS Genet. 2015, 11, e1005696. [Google Scholar] [CrossRef] [PubMed]
- Harkness, R.W.; Mittermaier, A.K. G-quadruplex dynamics. Biochim. Biophys. Acta-Proteins Proteomics 2017, 1865, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L. Four-stranded nucleic acids: Structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008, 37, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chemie Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Mergny, J.L.; Phan, A.T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Hurley, L.H.; Salazar, M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999, 27, 537–542. [Google Scholar] [CrossRef]
- Scalabrin, M.; Palumbo, M.; Richter, S.N. Highly Improved Electrospray Ionization-Mass Spectrometry Detection of G-Quadruplex-Folded Oligonucleotides and Their Complexes with Small Molecules. Anal. Chem. 2017, 89, 8632–8637. [Google Scholar] [CrossRef] [Green Version]
- Tretyakova, N.; Villalta, P.W.; Kotapati, S. Mass spectrometry of structurally modified DNA. Chem. Rev. 2013, 113, 2395–2436. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.; Sarkar, D.; Srivatsan, S.G. A Dual-App Nucleoside Probe Provides Structural Insights into the Human Telomeric Overhang in Live Cells. J. Am. Chem. Soc. 2018, 140, 12622–12633. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.-L.; Liu, H.; Xu, Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019, 47, 4940–4947. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cammas, A.; Millevoi, S. RNA G-quadruplexes: Emerging mechanisms in disease. Nucleic Acids Res. 2016, 45, gkw1280. [Google Scholar] [CrossRef] [PubMed]
- Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M.A.; Palù, G.; Flamand, L.; Calistri, A.; Richter, S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 2015, 118, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Ravichandran, S.; Kim, Y.-E.; Bansal, V.; Ghosh, A.; Hur, J.; Subramani, V.K.; Pradhan, S.; Lee, M.K.; Kim, K.K.; Ahn, J.-H. Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS Pathog. 2018, 14, e1007334. [Google Scholar] [CrossRef]
- Norseen, J.; Johnson, F.B.; Lieberman, P.M. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009, 83, 10336–10346. [Google Scholar] [CrossRef]
- Harris, L.M.; Monsell, K.R.; Noulin, F.; Famodimu, M.T.; Smargiasso, N.; Damblon, C.; Horrocks, P.; Merrick, C.J. G-Quadruplex DNA Motifs in the Malaria Parasite Plasmodium falciparum and Their Potential as Novel Antimalarial Drug Targets. Antimicrob. Agents Chemother. 2018, 62, e01828-17. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Alenko, A.; Burrows, C.J. Zika Virus Genomic RNA Possesses Conserved G-Quadruplexes Characteristic of the Flaviviridae Family. ACS Infect. Dis. 2016, 2, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.M.; Merrick, C.J. G-quadruplexes in pathogens: A common route to virulence control? PLoS Pathog. 2015, 11, e1004562. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palù, G.; Richter, S.N. A Dynamic G-Quadruplex Region Regulates the HIV-1 Long Terminal Repeat Promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef] [PubMed]
- Bhartiya, D.; Chawla, V.; Ghosh, S.; Shankar, R.; Kumar, N. Genome-wide regulatory dynamics of G-quadruplexes in human malaria parasite Plasmodium falciparum. Genomics 2016, 108, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Madireddy, A.; Purushothaman, P.; Loosbroock, C.P.; Robertson, E.S.; Schildkraut, C.L.; Verma, S.C. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 2016, 44, 3675–3694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artusi, S.; Perrone, R.; Lago, S.; Raffa, P.; Di Iorio, E.; Palù, G.; Richter, S.N. Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res. 2016, 44, 10343–10353. [Google Scholar] [CrossRef]
- Roizman, B.; Baines, J. The diversity and unity of herpesviridae. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 63–79. [Google Scholar] [CrossRef]
- Honess, R.W.; Roizman, B. Regulation of herpesvirus macromolecular synthesis: Sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 1975, 72, 1276–1280. [Google Scholar] [CrossRef]
- Honess, R.W.; Roizman, B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 1974, 14, 8–19. [Google Scholar]
- Babb, R.; Huang, C.C.; Aufiero, D.J.; Herr, W. DNA recognition by the herpes simplex virus transactivator VP16: A novel DNA-binding structure. Mol. Cell. Biol. 2001, 21, 4700–4712. [Google Scholar] [CrossRef] [PubMed]
- Callegaro, S.; Perrone, R.; Scalabrin, M.; Doria, F.; Palù, G.; Richter, S.N. A core extended naphtalene diimide G-quadruplex ligand potently inhibits herpes simplex virus 1 replication. Sci. Rep. 2017, 7, 2341. [Google Scholar] [CrossRef] [PubMed]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef] [PubMed]
- Pesola, J.M.; Zhu, J.; Knipe, D.M.; Coen, D.M. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J. Virol. 2005, 79, 14516–14525. [Google Scholar] [CrossRef] [PubMed]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef] [PubMed]
- Rice, S.A.; Knipe, D.M. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 1988, 62, 3814–3823. [Google Scholar] [PubMed]
- McGeoch, D.J.; Dolan, A.; Donald, S.; Rixon, F.J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 1985, 181, 1–13. [Google Scholar] [CrossRef]
- Dauber, B.; Saffran, H.A.; Smiley, J.R. The herpes simplex virus 1 virion host shutoff protein enhances translation of viral late mRNAs by preventing mRNA overload. J. Virol. 2014, 88, 9624–9632. [Google Scholar] [CrossRef]
- Lee, J.S.; Raja, P.; Knipe, D.M. Herpesviral ICP0 Protein Promotes Two Waves of Heterochromatin Removal on an Early Viral Promoter during Lytic Infection. MBio 2016, 7, e02007-15. [Google Scholar] [CrossRef]
- Kuddus, R.; Gu, B.; DeLuca, N.A. Relationship between TATA-binding protein and herpes simplex virus type 1 ICP4 DNA-binding sites in complex formation and repression of transcription. J. Virol. 1995, 69, 5568–5575. [Google Scholar] [Green Version]
- Chan, C.-Y.; Umar, M.I.; Kwok, C.K. Spectroscopic analysis reveals the effect of a single nucleotide bulge on G-quadruplex structures. Chem. Commun. (Camb.) 2019, 55, 2616–2619. [Google Scholar] [CrossRef] [PubMed]
- Dolan, A.; Jamieson, F.E.; Cunningham, C.; Barnett, B.C.; McGeoch, D.J. The genome sequence of herpes simplex virus type 2. J. Virol. 1998, 72, 2010–2021. [Google Scholar] [PubMed]
- Cohen, J.I. The Varicella-Zoster Virus Genome; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–14. [Google Scholar]
- Jones, J.O.; Sommer, M.; Stamatis, S.; Arvin, A.M. Mutational Analysis of the Varicella-Zoster Virus ORF62/63 Intergenic Region. J. Virol. 2006, 80, 3116–3121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norberg, P.; Tyler, S.; Severini, A.; Whitley, R.; Liljeqvist, J.-Å.; Bergström, T. A genome-wide comparative evolutionary analysis of herpes simplex virus type 1 and varicella zoster virus. PLoS ONE 2011, 6, e22527. [Google Scholar] [CrossRef] [PubMed]
- Szpara, M.L.; Gatherer, D.; Ochoa, A.; Greenbaum, B.; Dolan, A.; Bowden, R.J.; Enquist, L.W.; Legendre, M.; Davison, A.J. Evolution and diversity in human herpes simplex virus genomes. J. Virol. 2014, 88, 1209–1227. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Straus, S.E.; Ostrove, J.M. Directionality and further mapping of varicella zoster virus transcripts. Virus Res. 1988, 9, 249–261. [Google Scholar] [CrossRef]
- Che, X.; Zerboni, L.; Sommer, M.H.; Arvin, A.M. Varicella-zoster virus open reading frame 10 is a virulence determinant in skin cells but not in T cells in vivo. J. Virol. 2006, 80, 3238–3248. [Google Scholar] [CrossRef] [PubMed]
- Moriuchi, M.; Moriuchi, H.; Straus, S.E.; Cohen, J.I. Varicella-Zoster Virus (VZV) Virion-Associated Transactivator Open Reading Frame 62 Protein Enhances the Infectivity of VZV DNA. Virology 1994, 200, 297–300. [Google Scholar] [CrossRef]
- Moriuchi, H.; Moriuchi, M.; Straus, S.E.; Cohen, J.I. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 1993, 67, 2739–2746. [Google Scholar] [Green Version]
- Zhou, J.; Rosu, F.; Amrane, S.; Korkut, D.N.; Gabelica, V.; Mergny, J.L. Assembly of chemically modified G-rich sequences into tetramolecular DNA G-quadruplexes and higher order structures. Methods 2014, 67, 159–168. [Google Scholar] [CrossRef]
- Harrison, R.J.; Cuesta, J.; Chessari, G.; Martin, A.R.; Sanji, K.B.; Anthony, P.R.; Morrell, J.; Sharon, M.G.; Christopher, M.I.; Farial, A.T.; et al. Trisubstituted Acridine Derivatives as Potent and Selective Telomerase Inhibitors. J. Med. Chem. 2003, 46, 4463–4476. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L. Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Res. 2005, 33, e138. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Boutell, C.; Davido, D.J. HSV-1 ICP0: Paving the way for viral replication. Future Virol. 2011, 6, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Sedlackova, L.; Perkins, K.D.; Lengyel, J.; Strain, A.K.; van Santen, V.L.; Rice, S.A. Herpes simplex virus type 1 ICP27 regulates expression of a variant, secreted form of glycoprotein C by an intron retention mechanism. J. Virol. 2008, 82, 7443–7455. [Google Scholar] [CrossRef] [PubMed]
- Suk, H.; Knipe, D.M. Proteomic analysis of the herpes simplex virus 1 virion protein 16 transactivator protein in infected cells. Proteomics 2015, 15, 1957–1967. [Google Scholar] [PubMed] [Green Version]
- De Nicola, B.; Lech, C.J.; Heddi, B.; Regmi, S.; Frasson, I.; Perrone, R.; Richter, S.N.; Phan, A.T. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 2016, 44, 6442–6451. [Google Scholar] [CrossRef]
- Perrone, R.; Doria, F.; Butovskaya, E.; Frasson, I.; Botti, S.; Scalabrin, M.; Lago, S.; Grande, V.; Nadai, M.; Freccero, M.; et al. Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 2015, 58, 9639–9652. [Google Scholar] [CrossRef]
- Perrone, R.; Butovskaya, E.; Daelemans, D.; Palu, G.; Pannecouque, C.; Richter, S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014, 69, 3248–3258. [Google Scholar] [CrossRef] [Green Version]
- Harkness, J.M.; Kader, M.; DeLuca, N.A. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J. Virol. 2014, 88, 6847–6861. [Google Scholar] [CrossRef]
- Lopergolo, A.; Perrone, R.; Tortoreto, M.; Doria, F.; Beretta, G.L.; Zuco, V.; Freccero, M.; Borrello, M.G.; Lanzi, C.; Richter, S.N.; et al. Targeting of RET oncogene by naphthalene diimide-mediated gene promoter G-quadruplex stabilization exerts anti-tumor activity in oncogene-addicted human medullary thyroid cancer. Oncotarget 2016, 7, 49649–49663. [Google Scholar] [CrossRef] [Green Version]
- Tong, X.; Lan, W.; Zhang, X.; Wu, H.; Liu, M.; Cao, C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753–6763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, L.K.; Sale, J.E. Replication of G Quadruplex DNA. Genes (Basel) 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Tlučková, K.; Marušič, M.; Tóthová, P.; Bauer, L.; Šket, P.; Plavec, J.; Viglasky, V. Human Papillomavirus G-Quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, S.L.; Ebbinghaus, S.W.; Hurley, L.H. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009, 131, 10878–10891. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Liu, W.-J.; Guo, K.; Rusche, J.J.; Ebbinghaus, S.; Gokhale, V.; Hurley, L.H. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008, 7, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Rigo, R.; Sissi, C. Characterization of G4–G4 Crosstalk in the c-KIT Promoter Region. Biochemistry 2017, 56, 4309–4312. [Google Scholar] [CrossRef] [PubMed]
- Summers, B.C.; Leib, D.A. Herpes simplex virus type 1 origins of DNA replication play no role in the regulation of flanking promoters. J. Virol. 2002, 76, 7020–7029. [Google Scholar] [CrossRef]
- Zhi, Y.; Sandri-Goldin, R.M. Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J. Virol. 1999, 73, 3246–3257. [Google Scholar]
- Smith, I.L.; Hardwicke, M.A.; Sandri-Goldin, R.M. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 1992, 186, 74–86. [Google Scholar] [CrossRef]
- Sandri-Goldin, R.M. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol. 2011, 6, 1261–1277. [Google Scholar] [CrossRef]
- Sahakyan, A.B.; Chambers, V.S.; Marsico, G.; Santner, T.; Di Antonio, M.; Balasubramanian, S. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017, 7, 14535. [Google Scholar] [CrossRef] [PubMed]
- Omaga, C.A.; Fleming, A.M.; Burrows, C.J. The Fifth Domain in the G-Quadruplex-Forming Sequence of the Human NEIL3 Promoter Locks DNA Folding in Response to Oxidative Damage. Biochemistry 2018, 57, 2958–2970. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-C.; Feng, H.; Lin, Y.-C.; Guo, X.-R. New strategies against drug resistance to herpes simplex virus. Int. J. Oral Sci. 2016, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |




| HSV-1 | Leading Strand | Lagging Strand | C (%) |
|---|---|---|---|
| ICP0 | |||
| 1875 | GCGGGAGGGGCATGCTAATGGGGTTCTTTGGGG | 100 | |
| 1881 | GGGGCATGCTAATGGGGTTCTTTGGGGGACACCGGG | 100 | |
| 1966 | GGGGGCGCCGGGTTGGTCCCCGGGGACGGGG | 100 | |
| 2009 | GGGCCTGCCTCCCCTGGGACGCGCGGCCATTGGGGG | 100 | |
| 2066 | GGGGAGGGGAAAGGCGTGGGG | 100 | |
| ICP4 | |||
| R146532 | GGGCGGGGCGCGAGGGCGGGTGGG | 100 | |
| 146574 | GGGCGGGGCCGGGGGTTCGACCAACGGG | 100 | |
| 146578 | GGGGCCGGGGGTTCGACCAACGGGCCGCGGCCACGGG | 100 | |
| 146666 | GGGGTGGGCCCGCCGGGGGGGCGGGGGG | 100 | |
| 146947 | GGGGCCGGGGGTTCGACCAACGGGCCGCGGCCACGGG | 100 | |
| ICP22/ICP47 | |||
| 131746 | GGGCAGGGGGCGGGGCCCGGG | 100 | |
| R131803 | GGGCGGGACCGGGGGGCCCGGGGACGGCCAACGGGCGCGCGGGG | 100 | |
| 131857 | GGGACCAACGGGACGGCGGGCGGCCCAAGGG | 100 | |
| 132059 | GGGGGCGGGCCCGGGCGGCGGGGGGCGGGTCTCTCCGGCG | 100 | |
| 132065 | GGGCCCGGGCGGCGGGGGGCGGG | 100 | |
| 132070 | GGGCGGCGGGGGGCGGGTCTCTCCGGCG | 100 | |
| R132290 | GGGTGGGGTGGGCGGG | 100 | |
| 132411 | GGGGGCGGAGGAGGGGGGACGCGGGGGCGGAGGAGGGGG | 100 | |
| 132424 | GGGGGGACGCGGGGGCGGAGGAGGGGGGACGCGGGGG | 100 | |
| ICP27 | |||
| 113501 | GGGGCGGGGGCCCCGCCCGGGGGGCGG | 100 | |
| 113519 | GGGGGGCGGAACGAGGAGGGGTTTGGG | 100 |
| HSV-1 | HSV-2 | VZV | |
|---|---|---|---|
| ICP0 | ICP0 | ORF61 | |
| ICP4 | ICP4 | ORF62 | |
| IE genes/proteins | ICP27 | ICP27 | ORF4 |
| ICP47 | ICP47 | n.p. | |
| ICP22 | ICP22 | ORF63 |
| HSV-2 | Leading Strand | Lagging Strand | C (%) |
|---|---|---|---|
| ICP0 | |||
| 1571 | GGGAAGCCGGCGCGGGGCGGTCGCCGGGGCGGAGTCCGGG | 100 | |
| 1913 | GGGGGCGGGCACCACTCAGGGCCGCGCCGGCGGGGCGCCGGGGGG | 100 | |
| 2027 | GGGGACGGGGCCGCCCCGAGAGGGGGGG | 100 | |
| ICP4 | |||
| 149044 | GGGGCGCGCGGGGCGGGGGG | 100 | |
| 149088 | GGGGCCGGCGGGGGCCAACGGGAGCGCGGGG | 100 | |
| 149287 | GCGGACGCGCGGGCGTCGGGGCGGGG | 100 | |
| 149570 | GGGGCGGCAGTGGGGGGGGGTGG | 100 | |
| ICP22/ICP47 | |||
| 132299 | GGGGGGCCGGGCCGGGGGGACGGG | 100 | |
| 132325 | GGGGGGACGGGCCGGGGGGACGGG | 100 | |
| 132988 | GGGCCCGGACGGGGGGCGGG | 100 | |
| ICP27 | |||
| 114386 | GGGGACGGCGGGGGCGGGGGCGGTGACGCCCGACGGGGAGGG | 100 | |
| 114579 | GGGGCTGGGATGGCGGGTGTCCTCCGAGGGGG | 100 | |
| VZV | Leading Strand | Lagging Strand | C (%) |
| ORF61 | |||
| 104021 | GGGGTCCGCCGGGCGCCCAGAAACCGGGGGGGGGTTATTTTCGGGGGGGGG | 100 | |
| 105081 | GGGCGGGCGACGGGCGGG | 100 | |
| ORF62/ORF63 | |||
| 109246 | GGGGAGGAAATATGCGGTCGAGGGGGGGG | 99 | |
| 109703 | GCGGTTTTATGGGGTGTGGGCGGG | ||
| 110410 | GGGTAAAATGGCAATGGGGGATTCCGGGGCGGGAGACCTTCGATTGGG | 100 | |
| ORF4 | |||
| 4278 | GGGTGCAGGTAAGCTTGTTTGGGG | 100 |
| HSV-1 | Topology | Tm | Tm (B19) | ΔTm | TDS |
|---|---|---|---|---|---|
| ICP0 | |||||
| 1875 | parallel | 60.9 ± 0.1 | 73.3 ± 0.2 | 12.4 | + |
| 1881 | hybrid | 51.9 ± 0.3/45.9 ± 0.2 | 80.7 ± 1.2/66.3 ± 0.5 | 28.8/20.4 | + * |
| 1966 | parallel | 68.6 ± 0.1 | >90 | >21.4 | + |
| 2009 | hybrid | 54.9 ± 0.6/65.0 ± 0.4 | 69.2 ± 0.6/>90 | 14.3/>25 | +/− |
| 2066 | parallel | >90 | + | ||
| ICP4 | |||||
| R146532 | parallel | 79.5 ± 0.1 | >90 | >10.5 | + |
| 146574 | parallel | 63.4 ± 0.2 | >90 | >26.6 | + |
| 146578 | parallel | 66.2 ± 0.1 | 80.3 ± 0.4 | 14.1 | +/− |
| 146666 | parallel | >90 | + | ||
| 146947 | parallel | 69.2 ± 0.2 | >90 | >20.8 | + |
| ICP22/ICP47 | |||||
| 131746 | hybrid | >90/71.4 ± 0.8 | + | ||
| R131803 | hybrid | 73.7 ± 0.8/76.7 ± 0.9 | >90/85.1 ± 0.8 | >16.3/8.4 | + * |
| 131857 | hybrid | 62.4 ± 0.5/65.3 ± 0.2 | 69.6 ± 0.9/67.6 ± 0.4 | 7.2/2.3 | + * |
| 132059 | parallel | >90 | + | ||
| 132065 | hybrid | >90/78.8 ± 0.7 | + | ||
| 132070 | parallel | 74.2 ± 0.1 | >90 | >15.8 | + * |
| R132290 | parallel | >90 | + ** | ||
| 132411 | parallel | >90 | + | ||
| 132424 | parallel | >90 | + ** | ||
| ICP27 | |||||
| 113501 | parallel | >90 | +/− | ||
| 113519 | parallel | >90 | + | ||
| HSV-2 | |||||
| ICP0 | |||||
| 1571 | hybrid | 77.7 ± 0.8/79.0 ± 0.7 | >90/71.2 ± 1.1 | >12.3/−7.8 | +/− |
| 1913 | n.a. | ||||
| 2027 | parallel | >90 | + | ||
| ICP4 | |||||
| 149044 | parallel | >90 | + | ||
| 149088 | hybrid | 67.2 ± 0.7/72.6 ± 0.7 | 64.5 ± 0.9/>90 | −2.7/>17.4 | + |
| 149287 | parallel | 67.2 ± 0.7 | 84.1 ± 2.2 | 16.9 | + |
| 149570 | parallel | 81.8 ± 0.8 | >90 | >8.2 | + |
| ICP22/ICP47 | |||||
| 132299 | parallel | >90 | + | ||
| 132988 | parallel | >90 | + | ||
| 132325 | parallel | >90 | + | ||
| ICP27 | |||||
| 114386 | parallel | 75.9 ± 0.4 | >90 | >14.1 | + * |
| 114579 | parallel | 66.8 ± 0.9 | 83.4 ± 0.7 | 16.6 | + |
| VZV | |||||
| ORF61 | |||||
| 104021 | n.a. | ||||
| 105081 | parallel | 75.9 ± 0.4 | >90 | >14.1 | + |
| ORF62/ORF63 | |||||
| 109246 | parallel | >90 | + | ||
| 109703 | parallel | 57.7 ± 0.2 | 84.7 ± 0.5 | 27.0 | + |
| 110410 | parallel | 56.5 ± 0.4 | 85.2 ± 0.8 | 28.7 | +/− |
| ORF4 | |||||
| 4278 | hybrid | 57.5 ± 0.3/53.2 ± 0.5 | 79.2 ± 0.9/82.0 ± 0.9 | 21.7/28.8 | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frasson, I.; Nadai, M.; Richter, S.N. Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses. Molecules 2019, 24, 2375. https://doi.org/10.3390/molecules24132375
Frasson I, Nadai M, Richter SN. Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses. Molecules. 2019; 24(13):2375. https://doi.org/10.3390/molecules24132375
Chicago/Turabian StyleFrasson, Ilaria, Matteo Nadai, and Sara N. Richter. 2019. "Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses" Molecules 24, no. 13: 2375. https://doi.org/10.3390/molecules24132375
APA StyleFrasson, I., Nadai, M., & Richter, S. N. (2019). Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses. Molecules, 24(13), 2375. https://doi.org/10.3390/molecules24132375