Identification of Pyrvinium, an Anthelmintic Drug, as a Novel Anti-Adipogenic Compound Based on the Gene Expression Microarray and Connectivity Map
Abstract
1. Introduction
2. Results
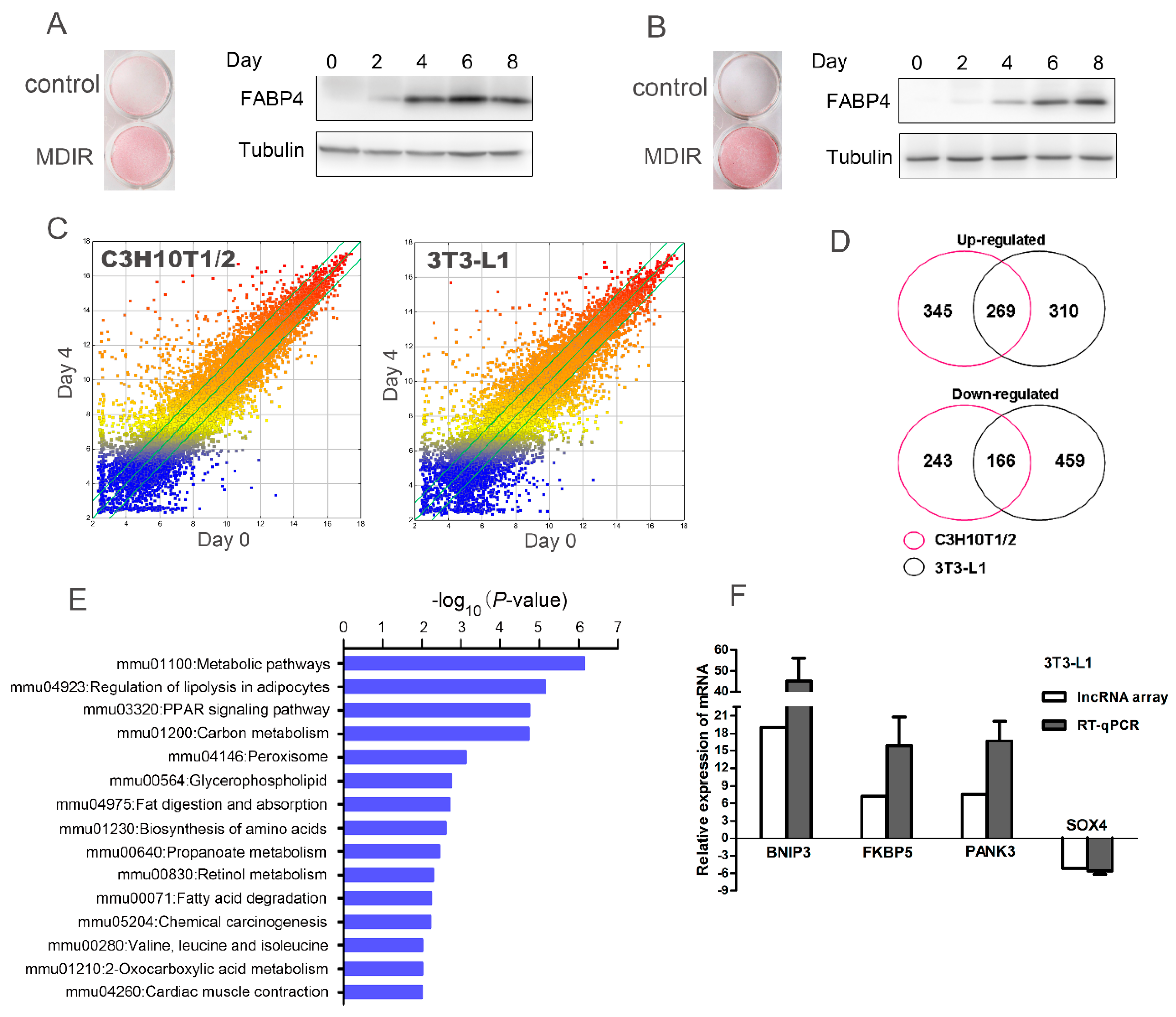
2.1. Microarray Analysis Identifies Differential Expression of mRNAs during Adipogenesis in C3H10T1/2 and 3T3-L1 Cells
2.2. Connectivity Map (C-Map) Analysis Identifies Pyrvinium as a Novel Anti-Adipogenic Compound
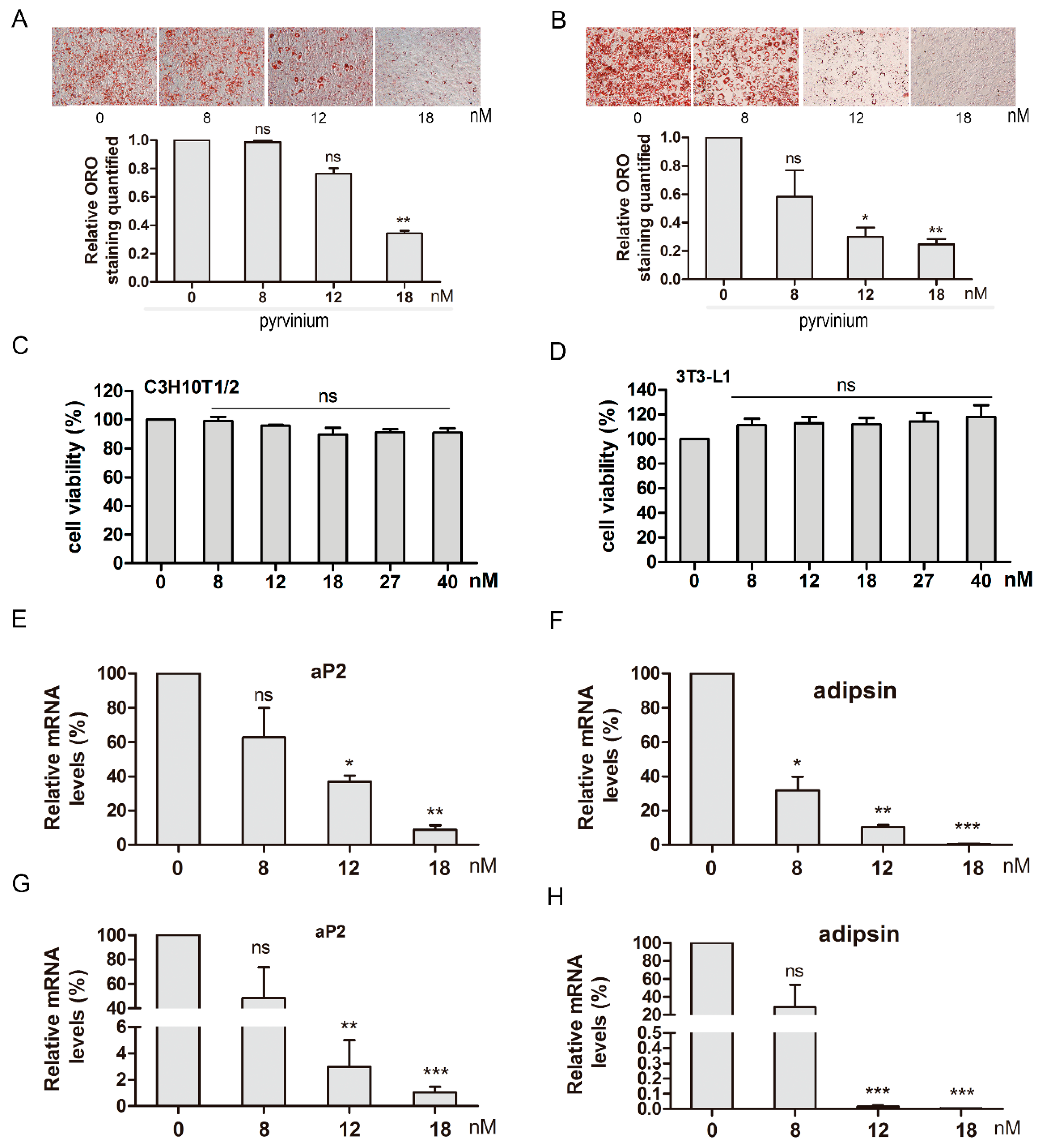
2.3. Pyrvinium Inhibits C3H10T1/2 and 3T3-L1 Adipogenesis in a Dose-Dependent Manner
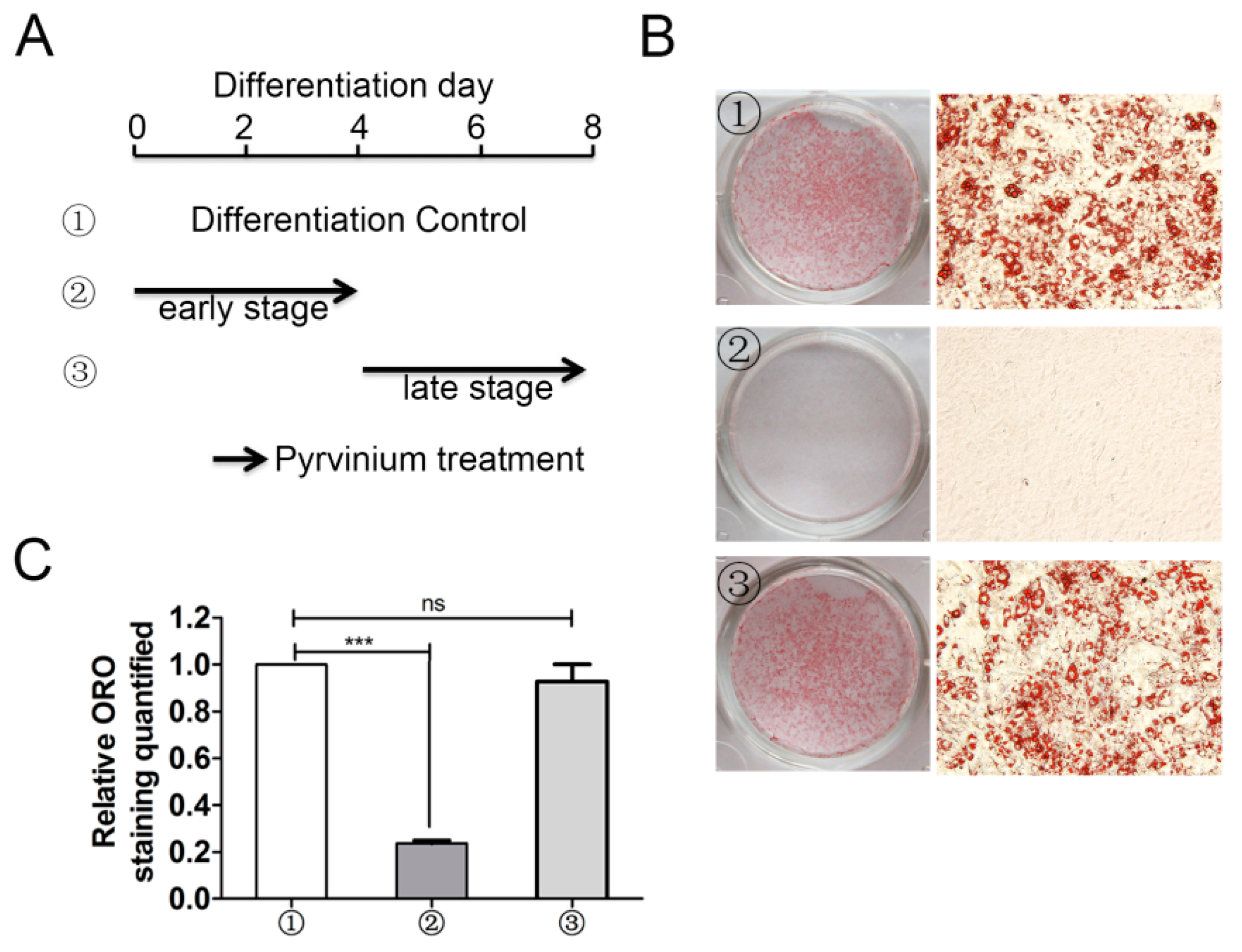
2.4. Early Stage Adipogenic Differentiation is Critical for the Inhibitory Action of Pyrvinium
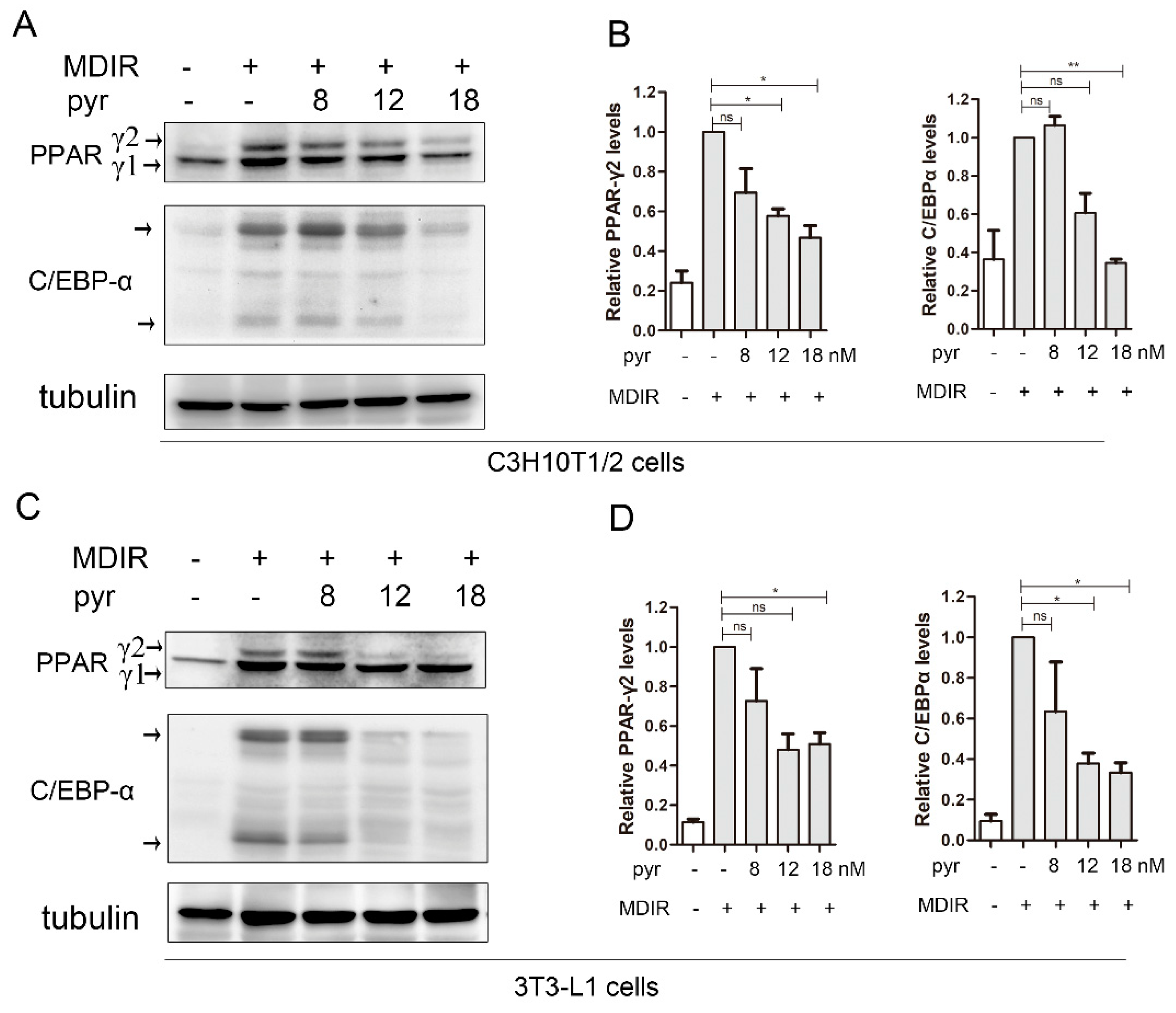
2.5. Pyrvinium Suppresses the Expression of Master Adipogenic Regulators
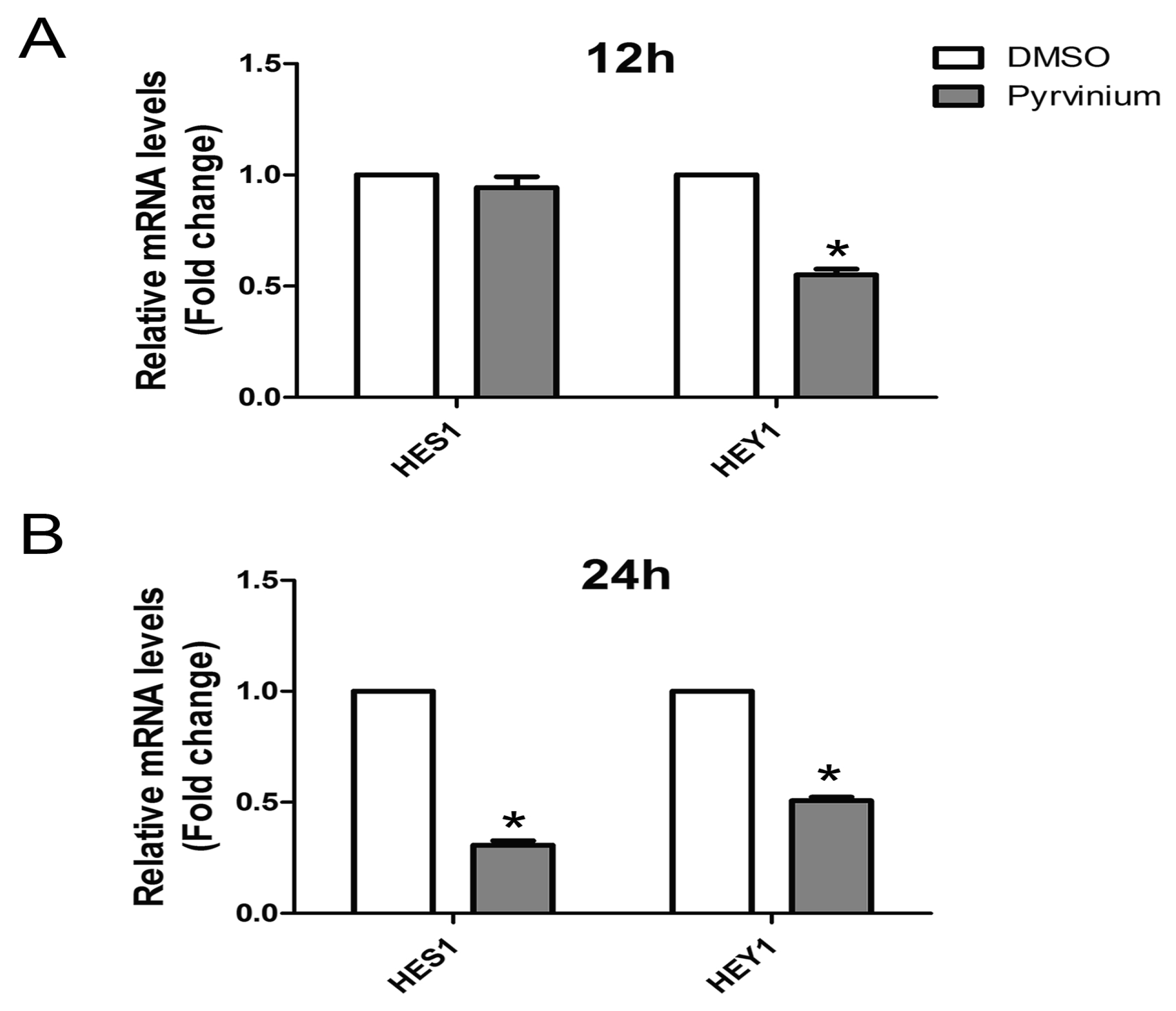
2.6. Effect of Pyrvinium on the Notch Signaling Pathway during Adipogenic Differentiation
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Adipogenic Differentiation
4.2. Oil Red O Staining
4.3. Microarray Gene Expression Studies and Connectivity Map (C-Map) Analysis
4.4. Cell Viability Assay
4.5. RNA Preparation and Real-Time PCR Analysis
4.6. Western Blot Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Haslam, D.W.; James, W.P.T. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From Stem Cell to Adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Boil. 2011, 12, 722–734. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Zhang, J.W.; Lane, M.D. Sequential gene promoter interactions by C/EBPβ, C/EBPα, and PPARγ during adipogenesis. Biochem. Biophys. Res. Commun. 2004, 318, 213–218. [Google Scholar] [CrossRef]
- Rangwala, S.M.; Lazar, M.A. T RANSCRIPTIONAL CONTROL OF A DIPOGENESIS. Annu. Rev. Nutr. 2000, 20, 535–559. [Google Scholar] [CrossRef]
- Zerbini, L.F.; Bhasin, M.K.; De Vasconcellos, J.F.; Paccez, J.D.; Gu, X.; Kung, A.L.; Libermann, T.A. Computational repositioning and preclinical validation of pentamidine for renal cell cancer. Mol. Cancer Ther. 2014, 13, 1929–1941. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Chen, X.; Zhang, Y.; Pan, X.; Wang, G.; Ye, Y. Expression Profiling Identifies Bezafibrate as Potential Therapeutic Drug for Lung Adenocarcinoma. J. Cancer 2015, 6, 1214–1221. [Google Scholar] [CrossRef]
- Cheng, H.W.; Liang, Y.H.; Kuo, Y.L.; Chuu, C.P.; Lin, C.Y.; Lee, M.H.; Wu, A.T.; Yeh, C.T.; Chen, E.I.; Whang-Peng, J.; et al. Identification of thioridazine, an antipsychotic drug, as an antiglioblastoma and anticancer stem cell agent using public gene expression data. Cell Death Dis. 2015, 6, e1753. [Google Scholar] [CrossRef]
- Guan, Q.; Jin, L.-J.; Chen, F.; Nie, Z.-Y. Gene Expression Profile and Functional Analysis of Alzheimer’s Disease. Am. J. Alzheimer’s Dis. Other Dementiasr 2013, 28, 693–701. [Google Scholar]
- Brum, A.M.; Van De Peppel, J.; Van Der Leije, C.S.; Schreuders-Koedam, M.; Eijken, M.; Van Der Eerden, B.C.J.; Van Leeuwen, J.P.T.M. Connectivity Map-based discovery of parbendazole reveals targetable human osteogenic pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 12711–12716. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lee, J.; Hernandez, M.A.S.; Mazitschek, R.; Ozcan, U. Treatment of Obesity with Celastrol. Cells 2015, 161, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Momtazi-Borojeni, A.A.; Abdollahi, E.; Ghasemi, F.; Caraglia, M.; Sahebkar, A. The novel role of pyrvinium in cancer therapy. J. Cell Physiol. 2018, 233, 2871–2881. [Google Scholar] [CrossRef] [PubMed]
- Carrella, D.; Manni, I.; Tumaini, B.; Dattilo, R.; Papaccio, F.; Mutarelli, M.; Sirci, F.; Amoreo, C.A.; Mottolese, M.; Iezzi, M.; et al. Computational drugs repositioning identifies inhibitors of oncogenic PI3K/AKT/P70S6K-dependent pathways among FDA-approved compounds. Oncotarget 2016, 7, 58743–58758. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhu, Y.; Lu, Y.; Yu, Z.; Zhong, J.; Li, Y.; Pan, J. Anthelmintic pyrvinium pamoate blocks Wnt/β-catenin and induces apoptosis in multiple myeloma cells. Oncol. Lett. 2018, 15, 5871–5878. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Zhang, L.; Zhou, Y.; Rajoria, P.; Wang, C. Pyrvinium selectively induces apoptosis of lymphoma cells through impairing mitochondrial functions and JAK2/STAT5. Biochem. Biophys. Res. Commun. 2016, 469, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fei, D.L.; Flaveny, C.A.; Dahmane, N.; Baubet, V.; Wang, Z.; Bai, F.; Pei, X.-H.; Rodriguez-Blanco, J.; Hang, B.; et al. Pyrvinium attenuates Hedgehog signaling downstream of Smoothened. Cancer Res. 2014, 74, 4811–4821. [Google Scholar] [CrossRef] [PubMed]
- Murakoshi, M.; Saiki, K.; Urayama, K.; Sato, T.N. An Anthelmintic Drug, Pyrvinium Pamoate, Thwarts Fibrosis and Ameliorates Myocardial Contractile Dysfunction in a Mouse Model of Myocardial Infarction. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Saraswati, S.; Deskins, D.L.; Holt, G.E.; Young, P.P. Pyrvinium, A Potent Small Molecule WNT Inhibitor, Increases Engraftment And Inhibits Lineage Commitment Of Mesenchymal Stem Cells (MSCs). Wound Repair Regen. 2012, 20, 185–193. [Google Scholar] [CrossRef]
- Desarzens, S.; Liao, W.-H.; Mammi, C.; Caprio, M.; Faresse, N. Hsp90 Blockers Inhibit Adipocyte Differentiation and Fat Mass Accumulation. PLoS ONE 2014, 9, e94127. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Z.; Zheng, R.D.; Wang, C.B.; Ying, Y.Q.; Luo, X.P. Effects of PI3K inhibitor LY294002 on the differentiation of mouse preadipocytes and the expression of C/EBPalpha and PPARgamma. Chin. J. Contemp. Pediatrics 2011, 13, 823–826. [Google Scholar]
- Dodson, M.V.; Hausman, G.J.; Poulos, S.P. Cell line models for differentiation: preadipocytes and adipocytes. Exp. Boil. Med. 2010, 235, 1185–1193. [Google Scholar]
- Jung, S.-R.; Song, N.-J.; Hwang, H.S.; An, J.J.; Cho, Y.-J.; Kweon, H.Y.; Kang, S.-W.; Lee, K.G.; Yoon, K.; Kim, B.-J.; et al. Silk peptides inhibit adipocyte differentiation through modulation of the Notch pathway in C3H10T1/2 cells. Nutr. Res. 2011, 31, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Ba, K.; Yang, X.; Wu, L.; Wei, X.; Fu, N.; Fu, Y.; Cai, X.; Yao, Y.; Ge, Y.; Lin, Y. Jagged-1-mediated activation of notch signalling induces adipogenesis of adipose-derived stem cells. Cell Prolif. 2012, 45, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Wang, M.; Wei, X.; Deng, S.; Fu, N.; Peng, Q.; Jiang, Y.; Ye, L.; Xie, J.; Lin, Y. Peroxisome Proliferator-Activated Receptor-gamma: Master Regulator of Adipogenesis and Obesity. Curr. Stem Cell Res. Ther. 2016, 11, 282–289. [Google Scholar] [CrossRef]
- De Sá, P.M.; Richard, A.J.; Hang, H.; Stephens, J.M. Transcriptional Regulation of Adipogenesis. Compr. Physiol. 2017, 272, 635–674. [Google Scholar]
- Lu, M.; Cao, Y.; Xiao, J.; Song, M.; Ho, C.-T. Molecular Mechanisms of the Anti-obesity Effect of Bioactive Ingredients in Common Spices: A Review. Food Funct. 2018, 9, 4569–4581. [Google Scholar] [CrossRef]
- Feng, S.; Reuss, L.; Wang, Y. Potential of Natural Products in the Inhibition of Adipogenesis through Regulation of PPARγ Expression and/or Its Transcriptional Activity. Molecules 2016, 21, 1278. [Google Scholar] [CrossRef]
- Moseti, D.; Regassa, A.; Kim, W.-K. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int. J. Mol. Sci. 2016, 17, 124. [Google Scholar] [CrossRef]
- Aranaz, P.; Navarro-Herrera, D.; Zabala, M.; Miguéliz, I.; Romo-Hualde, A.; López-Yoldi, M.; Martínez, J.A.; Vizmanos, J.L.; Milagro, F.I.; González-Navarro, C.J. Phenolic Compounds Inhibit 3T3-L1 Adipogenesis Depending on the Stage of Differentiation and Their Binding Affinity to PPARγ. Molecules 2019, 24, 1045. [Google Scholar] [CrossRef] [PubMed]
- Hadrich, F.; Sayadi, S. Apigetrin inhibits adipogenesis in 3T3-L1 cells by downregulating PPARγ and CEBP-α. Lipids Heal. Dis. 2018, 17, 95. [Google Scholar] [CrossRef] [PubMed]
- A Thorne, C.; Hanson, A.J.; Schneider, J.; Tahinci, E.; Orton, D.; Cselenyi, C.S.; Jernigan, K.K.; Meyers, K.C.; Hang, B.I.; Waterson, A.G.; et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat. Methods 2010, 6, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.E. Inhibition of Adipogenesis by Wnt Signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wang, J. Wnt/β-Catenin Signaling and Obesity. Front. Physiol. 2018, 9, 792. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef]
- Ross, D.A.; Rao, P.K.; Kadesch, T. Dual Roles for the Notch Target Gene Hes-1 in the Differentiation of 3T3-L1 Preadipocytes. Mol. Cell. Boil. 2004, 24, 3505–3513. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Dai, Z.; Pan, Y.; Wu, S.; Li, Z.; Zuo, C. E3 ubiquitin ligase DTX4 is required for adipogenic differentiation in 3T3-L1 preadipocytes cell line. Biochem. Biophys. Res. Commun. 2017, 492, 419–424. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Zhong, Y.; Zou, L.; Wang, Z.; Pan, Y.; Dai, Z.; Liu, X.; Cui, L.; Zuo, C. Lrrc75b is a novel negative regulator of C2C12 myogenic differentiation. Int. J. Mol. Med. 2016, 38, 1411–1418. [Google Scholar] [CrossRef][Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |
| Rank | C-Map Name | Mean C-Map Score 1 | N 2 | p Value |
|---|---|---|---|---|
| 1 | tanespimycin | −0.392 | 62 | <0.00001 |
| 2 | LY-294002 | −0.241 | 61 | 0.00002 |
| 3 | pyrvinium | −0.743 | 6 | 0.00048 |
| 4 | prochlorperazine | −0.424 | 16 | 0.00170 |
| 5 | terfenadine | −0.744 | 3 | 0.00172 |
| 6 | morantel | −0.587 | 5 | 0.00256 |
| 7 | triflusal | −0.806 | 3 | 0.00272 |
| 8 | thioridazine | −0.441 | 20 | 0.00552 |
| 9 | apigenin | −0.684 | 4 | 0.00577 |
| 10 | Methylbenzethonium chloride | −0.515 | 6 | 0.00683 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Dai, Z.; Luo, Z.; Zuo, C. Identification of Pyrvinium, an Anthelmintic Drug, as a Novel Anti-Adipogenic Compound Based on the Gene Expression Microarray and Connectivity Map. Molecules 2019, 24, 2391. https://doi.org/10.3390/molecules24132391
Wang Z, Dai Z, Luo Z, Zuo C. Identification of Pyrvinium, an Anthelmintic Drug, as a Novel Anti-Adipogenic Compound Based on the Gene Expression Microarray and Connectivity Map. Molecules. 2019; 24(13):2391. https://doi.org/10.3390/molecules24132391
Chicago/Turabian StyleWang, Zonggui, Zhong Dai, Zhicong Luo, and Changqing Zuo. 2019. "Identification of Pyrvinium, an Anthelmintic Drug, as a Novel Anti-Adipogenic Compound Based on the Gene Expression Microarray and Connectivity Map" Molecules 24, no. 13: 2391. https://doi.org/10.3390/molecules24132391
APA StyleWang, Z., Dai, Z., Luo, Z., & Zuo, C. (2019). Identification of Pyrvinium, an Anthelmintic Drug, as a Novel Anti-Adipogenic Compound Based on the Gene Expression Microarray and Connectivity Map. Molecules, 24(13), 2391. https://doi.org/10.3390/molecules24132391






