Experimental Insight into the Structural and Functional Roles of the ‘Black’ and ‘Gray’ Clusters in Recoverin, a Calcium Binding Protein with Four EF-Hand Motifs
Abstract
:1. Introduction
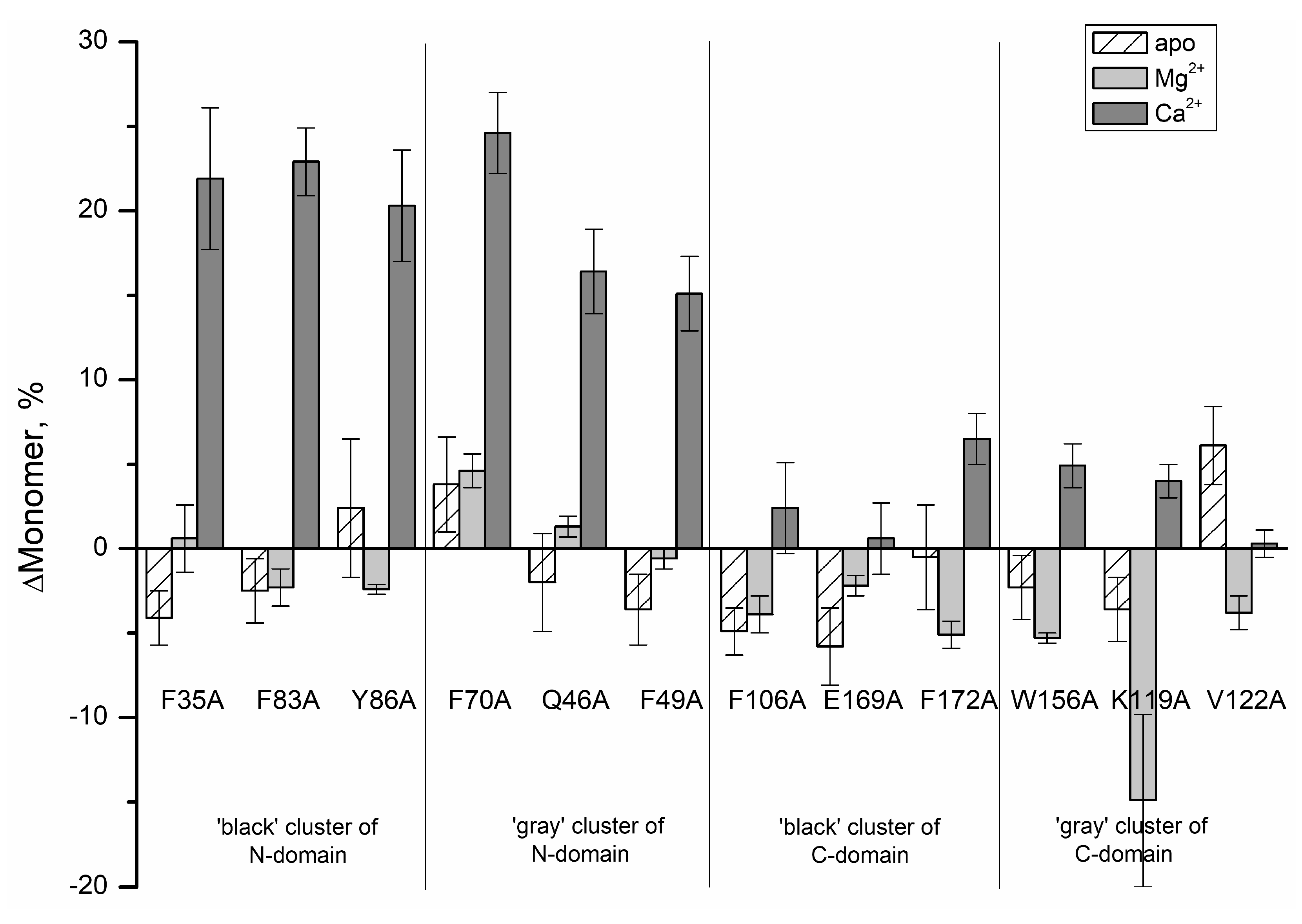
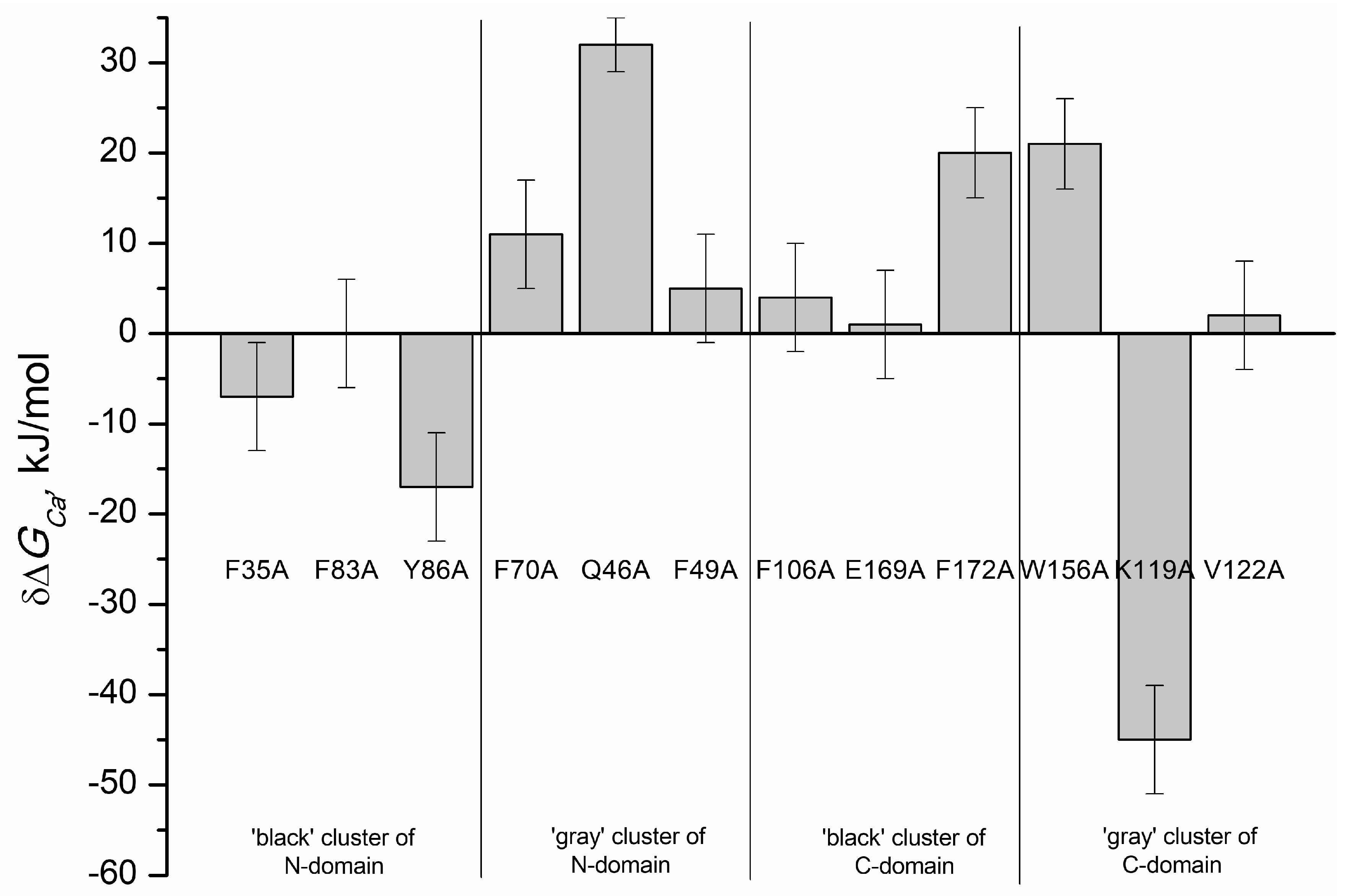
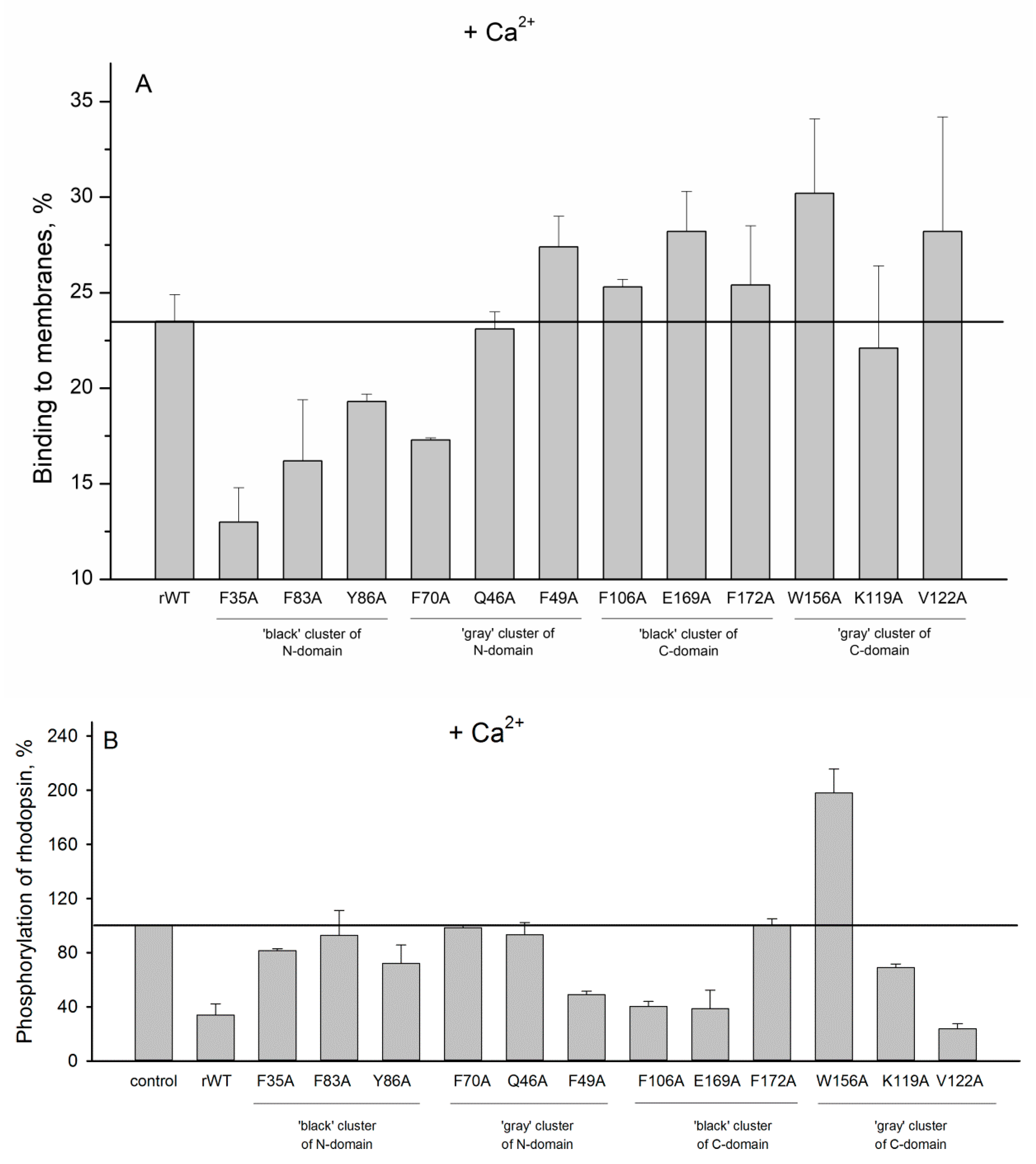
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Expression and Purification of Human Wild-Type Recoverin and Its Mutants
3.3. Removal of Metal Ions from Recoverin
3.4. Chemical Crosslinking of Recoverin and Its Mutants
3.5. Circular Dichroism Measurements
3.6. Fluorescence Measurements
3.7. Evaluation of Calcium Binding Parameters by Means of The Calcium Buffer Method
α = 1/(1 + f1 + f1 × f2),
f1 = 10(pK1 - pCa),
f2 = 10(pK2 - pCa).
f = 10[(logK − pM) × n].
3.8. Determination of Metal Affinity of Recoverin from Protein Titration by Ca2+
P + M ↔ P∙M,
Ka2
P∙M + M ↔ P∙M2,
Protein + M ↔ Protein∙M,
3.9. The Equilibrium Centrifugation Assay of Recoverin Binding to Membranes
3.10. Rhodopsin Phosphorylation Assay
3.11. Computational Evaluation of the Intrinsic Disorder Predisposition of Human Recoverin and Its Cluster Mutants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Permyakov, E.A. Parvalbumin; Nova Science Publishers: New York, NY, USA, 2006. [Google Scholar]
- Permyakov, E.A. Metalloproteomics; Uversky, V.N., Ed.; Wiley Series in Protein and Peptide Science; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009. [Google Scholar]
- Permyakov, E.A.; Kretsinger, R.H. Calcium Binding Proteins.; Uversky, V.N., Ed.; Wiley Series in protein and Peptide Science; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Nockolds, C.E.; Kretsinger, R.H.; Coffee, C.J.; Bradshaw, R.A. Structure of a calcium-binding carp myogen. Proc. Natl. Acad. Sci. USA 1972, 69, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Kretsinger, R.H.; Nockolds, C.E. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 1973, 248, 3313–3326. [Google Scholar] [PubMed]
- Moews, P.C.; Kretsinger, R.H. Refinement of the structure of carp muscle calcium-binding parvalbumin by model building and difference Fourier analysis. J. Biol. Chem. 1975, 91, 201–225. [Google Scholar] [CrossRef]
- Denessiouk, K.; Permyakov, S.; Denesyuk, A.; Permyakov, E.; Johnson, M.S. Two structural motifs within canonical EF-hand calcium binding domains identify five different classes of calcium buffers and sensors. PLoS ONE 2014, 9, e109287. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Vologzhannikova, A.A.; Khorn, P.A.; Shevelyova, M.P.; Kazakov, A.S.; Emelyanenko, V.I.; Denesyuk, A.I.; Denessiouk, K.; Uversky, V.N.; Permyakov, E.A. Comprehensive analysis of the roles of ’black’ and ’gray’ clusters in structure and function of rat beta-parvalbumin. Cell Calcium 2018, 75, 64–78. [Google Scholar] [CrossRef]
- Permyakova, M.E.; Permyakov, S.E.; Kazakov, A.S.; Denesyuk, A.I.; Denessiouk, K.; Uversky, V.N.; Permyakov, E.A. Analyzing the structural and functional roles of residues from the ’black’ and ’gray’ clusters of human S100P protein. Cell Calcium 2019, 80, 46–55. [Google Scholar] [CrossRef]
- Burgoyne, R.D.; O’Callaghan, D.W.; Hasdemir, B.; Haynes, L.P.; Tepikin, A.V. Neuronal Ca2+-sensor proteins: Multitalented regulators of neuronal function. Trends Neurosci. 2004, 27, 203–209. [Google Scholar] [CrossRef]
- Ames, J.B.; Ishima, R.; Tanaka, T.; Gordon, J.I.; Stryer, L.; Ikura, M. Molecular mechanics of calcium-myristoyl switches. Nature 1997, 389, 198–202. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Ericsson, L.H.; Johnson, R.S.; Kumar, S.; Olshevskaya, E.; Zozulya, S.; Neubert, T.A.; Stryer, L.; Hurley, J.B.; Walsh, K.A. The NH2 terminus of retinal recoverin is acylated by a small family of fatty acids. J. Biol. Chem. 1992, 267, 16033–16036. [Google Scholar]
- Tanaka, T.; Ames, J.B.; Harvey, T.S.; Stryer, L.; Ikura, M. Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 1995, 376, 444–447. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Cherskaya, A.M.; Senin, I.I.; Zargarov, A.A.; Shulga-Morskoy, S.V.; Alekseev, A.M.; Zinchenko, D.V.; Lipkin, V.M.; Philippov, P.P.; Uversky, V.N.; et al. Effects of mutations in the calcium-binding sites of recoverin on its calcium affinity: Evidence for successive filling of the calcium binding sites. Protein Eng. 2000, 13, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Zozulya, S.; Stryer, L. Calcium-myristoyl protein switch. Proc. Natl. Acad Sci. USA 1992, 89, 11569–11573. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B.; Tanaka, T.; Ikura, M.; Stryer, L. Nuclear magnetic resonance evidence for Ca2+-induced extrusion of the myristoyl group of recoverin. J. Biol. Chem. 1995, 270, 30909–30913. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B.; Ikura, M. Structure and membrane-targeting mechanism of retinal Ca2+-binding proteins, recoverin and GCAP-2. Adv. Exp. Med. Biol. 2002, 514, 333–348. [Google Scholar] [PubMed]
- Ames, J.B.; Hamasaki, N.; Molchanova, T. Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state. Biochemistry 2002, 41, 5776–5787. [Google Scholar] [CrossRef] [PubMed]
- Senin, I.I.; Fischer, T.; Komolov, K.E.; Zinchenko, D.V.; Philippov, P.P.; Koch, K.W. Ca2+-myristoyl switch in the neuronal calcium sensor recoverin requires different functions of Ca2+-binding sites. J. Biol. Chem. 2002, 277, 50365–50372. [Google Scholar] [CrossRef]
- Lange, C.; Koch, K.W. Calcium-dependent binding of recoverin to membranes monitored by surface plasmon resonance spectroscopy in real time. Biochemistry 1997, 36, 12019–12026. [Google Scholar] [CrossRef]
- Kawamura, S.; Hisatomi, O.; Kayada, S.; Tokunaga, F.; Kuo, C.H. Recoverin has S-modulin activity in frog rods. J. Biol. Chem. 1993, 268, 14579–14582. [Google Scholar]
- Gorodovikova, E.N.; Senin, I.I.; Philippov, P.P. Calcium-sensitive control of rhodopsin phosphorylation in the reconstituted system consisting of photoreceptor membranes, rhodopsin kinase and recoverin. FEBS Lett 1994, 353, 171–172. [Google Scholar] [CrossRef] [Green Version]
- Erickson, M.A.; Lagnado, L.; Zozulya, S.; Neubert, T.A.; Stryer, L.; Baylor, D.A. The effect of recombinant recoverin on the photoresponse of truncated rod photoreceptors. Proc. Natl. Acad Sci. USA 1998, 95, 6474–6479. [Google Scholar] [CrossRef] [Green Version]
- Burns, M.E.; Baylor, D.A. Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu. Rev. Neurosci. 2001, 24, 779–805. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, E.A. Luminescent Spectroscopy of Proteins; CRC Press, Inc.: Boca Raton, FL, USA; Ann Arbor, MI, USA; London, UK; Tokyo, Japan, 1993. [Google Scholar]
- Landau, L.D.; Lifshitz, L.P. Statistical Physics, Part. 1., 3rd ed.; Pergamon Press: Oxford, UK, 1980. [Google Scholar]
- Ames, J.B.; Lim, S. Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta 2012, 1820, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B.; Levay, K.; Wingard, J.N.; Lusin, J.D.; Slepak, V.Z. Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J. Biol. Chem. 2006, 281, 37237–37245. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Bakunts, A.G.; Permyakova, M.E.; Denesyuk, A.I.; Uversky, V.N.; Permyakov, E.A. Metal-controlled interdomain cooperativity in parvalbumins. Cell Calcium. 2009, 46, 163–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Permyakov, S.E.; Nazipova, A.A.; Denesyuk, A.I.; Bakunts, A.G.; Zinchenko, D.V.; Lipkin, V.M.; Uversky, V.N.; Permyakov, E.A. Recoverin as a redox-sensitive protein. J. Proteome Res. 2007, 6, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Zernii, E.Y.; Komolov, K.E.; Permyakov, S.E.; Kolpakova, T.; Dell’orco, D.; Poetzsch, A.; Knyazeva, E.L.; Grigoriev, I.I.; Permyakov, E.A.; Senin, I.I.; et al. Involvement of the recoverin C-terminal segment in recognition of the target enzyme rhodopsin kinase. Biochem. J. 2011, 435, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laible, M.; Boonrod, K. Homemade site directed mutagenesis of whole plasmids. J. Vis. Exp. 2009, 27, 1135. [Google Scholar] [CrossRef]
- Blum, H.E.; Lehky, P.; Kohler, L.; Stein, E.A.; Fischer, E.H. Comparative properties of vertebrate parvalbumins. J. Biol. Chem. 1977, 252, 2834–2838. [Google Scholar] [PubMed]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 2000, 287, 243–251. [Google Scholar] [CrossRef]
- Schwarzenbach, G.; Flaschka, H. Die Komplexometrische Titration; Ferdinand Enke Verlag: Stuttgart, Germany, 1965. [Google Scholar]
- Permyakov, S.E.; Zernii, E.Y.; Knyazeva, E.L.; Denesyuk, A.I.; Nazipova, A.A.; Kolpakova, T.V.; Zinchenko, D.V.; Philippov, P.P.; Permyakov, E.A.; Senin, I.I. Oxidation mimicking substitution of conservative cysteine in recoverin suppresses its membrane association. Amino Acids 2012, 42, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Vologzhannikova, A.A.; Khorn, P.A.; Kazakov, A.S.; Ismailov, R.G.; Sokolov, A.S.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. In search for globally disordered apo-parvalbumins: Case of parvalbumin beta-1 from coho salmon. Cell Calcium 2017, 67, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.I.; Senin, I.I.; Tikhomirova, N.K.; Komolov, K.E.; Permyakov, S.E.; Zernii, E.Y.; Koch, K.W.; Philippov, P.P. Synergetic effect of recoverin and calmodulin on regulation of rhodopsin kinase. Front Mol. Neurosci. 2012, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Cherskaya, A.M.; Wasserman, L.A.; Khokhlova, T.I.; Senin, I.I.; Zargarov, A.A.; Zinchenko, D.V.; Zernii, E.Y.; Lipkin, V.M.; Philippov, P.P.; et al. Recoverin is a zinc-binding protein. J. Proteome Res. 2003, 2, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.O.; Roman, A.Y.; Baksheeva, V.E.; Nazipova, A.A.; Shevelyova, M.P.; Vladimirov, V.I.; Buyanova, M.F.; Zinchenko, D.V.; Zamyatnin, A.A., Jr.; Devred, F.; et al. Functional status of neuronal calcium sensor-1 is modulated by zinc binding. Front Mol. Neurosci. 2018, 11, 459. [Google Scholar] [CrossRef]
- Senin, I.I.; Tikhomirova, N.K.; Churumova, V.A.; Grigoriev, I.I.; Kolpakova, T.A.; Zinchenko, D.V.; Philippov, P.P.; Zernii, E.Y. Amino acid sequences of two immune-dominant epitopes of recoverin are involved in Ca2+/recoverin-dependent inhibition of phosphorylation of rhodopsin. Biochemistry-Moscow 2011, 76, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Zernii, E.Y.; Nazipova, A.A.; Nemashkalova, E.L.; Kazakov, A.S.; Gancharova, O.S.; Serebryakova, M.V.; Tikhomirova, N.K.; Baksheeva, V.E.; Vladimirov, V.I.; Zinchenko, D.V.; et al. Light-induced thiol oxidation of recoverin affects rhodopsin desensitization. Front Mol. Neurosci. 2018, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Vladimirov, V.I.; Zernii, E.Y.; Baksheeva, V.E.; Wimberg, H.; Kazakov, A.S.; Tikhomirova, N.K.; Nemashkalova, E.L.; Mitkevich, V.A.; Zamyatnin, A.A., Jr.; Lipkin, V.M.; et al. Photoreceptor calcium sensor proteins in detergent-resistant membrane rafts are regulated via binding to caveolin-1. Cell Calcium 2018, 73, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Peng, Z.L.; Kurgan, L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012, 13, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Kurgan, L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J. Biomol. Struct. Dyn. 2014, 32, 448–464. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Silman, I.; Sussman, J.L. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: An overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Kurgan, L. On the complementarity of the consensus-based disorder prediction. Pac. Symp. Biocomput. 2012, 176–187. [Google Scholar]
- Meng, F.; Uversky, V.; Kurgan, L. Computational prediction of intrinsic disorder in proteins. Curr. Protoc. Protein Sci. 2017, 88, 2.16.1–2.16.14. [Google Scholar]
- Walsh, I.; Giollo, M.; Di Domenico, T.; Ferrari, C.; Zimmermann, O.; Tosatto, S.C. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics 2015, 31, 201–208. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds wild type recombinant recoverin and its mutants are available from the authors. |









| Protein | Protein State | α-Helices % | β-Sheets % | Turns % | Unordered Structure | ||
|---|---|---|---|---|---|---|---|
| rWT | Apo-form | 48.1 ± 0.1 | 8.9 ± 0.3 | 16.5 ± 0.2 | 26.8 ± 0.3 | ||
| N-domain | ‘black’ cluster | F35A | 47.9 ± 0.2 | 8.6 ± 0.4 | 16.6 ± 0.2 | 26.7 ± 0.3 | |
| F83A | 49.3 ± 0.3 | 7.5 ± 0.3 | 15.9 ± 0.2 | 27.0 ± 0.1 | |||
| Y86A | 48.5 ± 0.2 | 8.0 ± 0.2 | 15.7 ± 0.2 | 27.9 ± 0.3 | |||
| ‘gray’ cluster | F70A | 47.9 ± 0.3 | 8.4 ± 0.3 | 16.1 ± 0.3 | 27.6 ± 0.3 | ||
| Q46A | 48.1 ± 0.4 | 7.9 ± 0.2 | 15.7 ± 0.2 | 27.8 ± 0.1 | |||
| F49A | 46.4 ± 0.5 | 8.9 ± 0.3 | 16.4 ± 0.4 | 28.5 ± 0.5 | |||
| C-domain | ‘black’ cluster | F106A | 49.7 ± 0.1 | 7.6 ± 0.4 | 15.8 ± 0.1 | 26.8 ± 0.4 | |
| E169A | 46.9 ± 0.9 | 8.7 ± 0.3 | 17.4 ± 0.2 | 27.3 ± 0.7 | |||
| F172A | 50.0 ± 0.3 | 7.9 ± 0.2 | 16.4 ± 0.4 | 25.3 ± 0.3 | |||
| ‘gray’ cluster | W156A | 47.0 ± 0.3 | 9.0 ± 0.3 | 17.0 ± 0.1 | 27.4 ± 0.1 | ||
| K119A | 49.2 ± 0.3 | 8.1 ± 0.3 | 16.1 ± 0.2 | 26.4 ± 0.2 | |||
| V122A | 47.1 ± 0.2 | 8.9 ± 0.3 | 16.4 ± 0.1 | 27.9 ± 0.2 | |||
| rWT | Mg2+ | 49.3 ± 0.1 | 7.9 ± 0.3 | 17.9 ± 0.2 | 25.3 ± 0.3 | ||
| N-domain | ‘black’ cluster | F35A | 49.9 ± 0.2 | 8.2 ± 0.4 | 16.8 ± 0.2 | 25.4 ± 0.3 | |
| F83A | 51.1 ± 0.3 | 7.1 ± 0.3 | 15.9 ± 0.2 | 26.0 ± 0.1 | |||
| Y86A | 50.9 ± 0.2 | 7.3 ± 0.2 | 16.6 ± 0.2 | 25.3 ± 0.3 | |||
| N-domain | ‘gray’ cluster | F70A | 48.2 ± 0.3 | 8.0 ± 0.3 | 17.4 ± 0.3 | 26.6 ± 0.3 | |
| Q46A | 50.9 ± 0.4 | 7.8 ± 0.2 | 16.0 ± 0.2 | 25.3 ± 0.1 | |||
| F49A | 48.4 ± 0.5 | 7.9 ± 0.3 | 16.8 ± 0.4 | 27.1 ± 0.5 | |||
| C-domain | ‘black’ cluster | F106A | 53.2 ± 0.1 | 7.0 ± 0.4 | 16.2 ± 0.1 | 24.2 ± 0.4 | |
| E169A | 49.8 ± 0.9 | 7.4 ± 0.3 | 17.1 ± 0.2 | 26.2 ± 0.7 | |||
| F172A | 51.1 ± 0.3 | 7.9 ± 0.2 | 15.7 ± 0.4 | 25.5 ± 0.3 | |||
| ‘gray’ cluster | W156A | 49.1 ± 0.3 | 7.4 ± 0.3 | 17.4 ± 0.1 | 26.3 ± 0.1 | ||
| K119A | 51.3 ± 0.3 | 7.8 ± 0.3 | 15.5 ± 0.2 | 25.4 ± 0.2 | |||
| V122A | 48.9 ± 0.2 | 7.9 ± 0.3 | 16.7 ± 0.1 | 26.5 ± 0.2 | |||
| rWT | Ca2+ | 51.2 ± 0.1 | 7.0 ± 0.3 | 16.0 ± 0.2 | 25.7 ± 0.3 | ||
| N-domain | ‘black’ cluster | F35A | 55.0 ± 0.2 | 7.5 ± 0.4 | 14.3 ± 0.2 | 23.5 ± 0.3 | |
| F83A | 57.4 ± 0.3 | 5.4 ± 0.3 | 14.4 ± 0.2 | 22.5 ± 0.1 | |||
| Y86A | 56.9 ± 0.2 | 5.4 ± 0.2 | 13.9 ± 0.2 | 23.5 ± 0.3 | |||
| ‘gray’ cluster | F70A | 52.0 ± 0.3 | 7.0 ± 0.3 | 15.8 ± 0.3 | 25.1 ± 0.3 | ||
| Q46A | 54.6 ± 0.4 | 7.0 ± 0.2 | 15.4 ± 0.2 | 23.0 ± 0.1 | |||
| F49A | 53.9 ± 0.5 | 6.9 ± 0.3 | 15.9 ± 0,4 | 22.8 ± 0.5 | |||
| C-domain | ‘black’ cluster | F106A | 55.7 ± 0.1 | 5.8 ± 0.4 | 14.0 ± 0.1 | 24.2 ± 0.4 | |
| E169A | 52.3 ± 0.9 | 6.8 ± 0.3 | 15.3 ± 0.2 | 25.3 ± 0.7 | |||
| F172A | 54.2 ± 0.3 | 6.6 ± 0.2 | 15.5 ± 0.4 | 23.4 ± 0.3 | |||
| ‘gray’ cluster | W156A | 52.4 ± 0.3 | 7.1 ± 0.3 | 15.5 ± 0.1 | 24.9 ± 0.1 | ||
| K119A | 54.4 ± 0.3 | 6.8 ± 0.3 | 15.4 ± 0.2 | 23.5 ± 0.2 | |||
| V122A | 57.6 ± 0.2 | 6.0 ± 0.3 | 14.4 ± 0.1 | 22.0 ± 0.2 | |||
| Protein | Protein State | Monomer 20 kDa, % | Dimer 50 kDa, % | Trimer 70–100 kDa, % | Multimer 100–250 kDa, % | ||
|---|---|---|---|---|---|---|---|
| rWT | Apo-state | 66.9 ± 1.1 | 33.1 ± 1.1 | - | - | ||
| N-domain | ‘black’ cluster | F35A | 62.8 ± 0.5 | 37.2 ± 0.5 | - | - | |
| F83A | 64.4 ± 0.8 | 35.6 ± 0.8 | - | - | |||
| Y86A | 69.3 ± 3.0 | 30.7 ± 3.0 | - | - | |||
| ‘gray’ cluster | F70A | 70.7 ± 1.7 | 29.3 ± 1.7 | - | - | ||
| Q46A | 64.9 ± 1.8 | 35.1 ± 1.8 | - | - | |||
| F49A | 63.3 ± 1.0 | 36.7 ± 1.0 | - | - | |||
| C-domain | ‘black’ cluster | F106A | 62.0 ± 0.3 | 38.0 ± 0.3 | - | - | |
| E169A | 61.1 ± 1.2 | 38.9 ± 1.2 | - | - | |||
| F172A | 66.4 ± 2.0 | 33.6 ± 2.0 | - | - | |||
| ‘gray’ cluster | W156A | 64.6 ± 0.8 | 35.4 ± 0.8 | - | - | ||
| K119A | 63.3 ± 0.8 | 36.7 ± 0.8 | - | - | |||
| V122A | 73.0 ± 1.2 | 27.0 ± 1.2 | - | - | |||
| rWT | Mg2+-loaded | 68.45 ± 0.09 | 31.55 ± 0.09 | - | - | ||
| N-domain | ‘black’ cluster | F35A | 69.1 ± 1.9 | 30.9 ± 1.9 | - | ||
| F83A | 66.2 ± 1.0 | 33.8 ± 1.0 | - | ||||
| Y86A | 66.1 ± 0.2 | 33.9 ± 0.2 | - | ||||
| ‘gray’ cluster | F70A | 73.1 ± 0.9 | 26.9 ± 0.9 | - | - | ||
| Q46A | 69.8 ± 0.5 | 30.2 ± 0.5 | - | - | |||
| F49A | 67.9 ± 0.5 | 32.1 ± 0.5 | - | - | |||
| C-domain | ‘black’ cluster | F106A | 64.6 ± 1.0 | 35.4 ± 1.0 | - | - | |
| E169A | 66.3 ± 0.5 | 33.7 ± 0.5 | - | - | |||
| F172A | 63.4 ± 0.7 | 36.6 ± 0.7 | - | - | |||
| ‘gray’ cluster | W156A | 63.2 ± 0.2 | 36.8 ± 0.2 | - | - | ||
| K119A | 53.6 ± 5.0 | 46.4 ± 5.0 | - | - | |||
| V122A | 64.7 ± 0.9 | 35.3 ± 0.9 | - | - | |||
| rWT | Ca2+-loaded | 15.5 ± 0.6 | 30.9 ± 0.4 | 32.0 ± 1.1 | 21.55 ± 0.09 | ||
| N- domain | ‘black’ cluster | F35A | 37.4 ± 3.6 | 31.7 ± 0.3 | 17.78 ± 0.07 | 13.2 ± 3.3 | |
| F83A | 38.4 ± 1.4 | 28.7 ± 0.8 | 19.7 ± 0.4 | 13.2 ± 0.3 | |||
| Y86A | 35.8 ± 2.7 | 28.5 ± 1.4 | 21.5 ± 0.7 | 14.2 ± 0.6 | |||
| ‘gray’ cluster | F70A | 40.1 ± 1.8 | 29.3 ± 1.1 | 19.1 ± 0.5 | 11.4 ± 0.2 | ||
| Q46A | 31.9 ± 1.9 | 27.3 ± 1.5 | 22.3 ± 1.0 | 18.4 ± 0.6 | |||
| F49A | 30.6 ± 1.6 | 25.5 ± 0.5 | 22.8 ± 1.3 | 21.08 ± 0.09 | |||
| C-domain | ‘black’ cluster | F106A | 17.9 ± 2.1 | 27.11 ± 0.08 | 30.0 ± 1.4 | 25.0 ± 0.8 | |
| E169A | 16.1 ± 1.5 | 27.0 ± 0.3 | 29.6 ± 0.3 | 27.2 ± 0.9 | |||
| F172A | 22.0 ± 0.9 | 29.3 ± 0.2 | 28.8 ± 0.8 | 20.0 ± 0.3 | |||
| ‘gray’ cluster | W156A | 20.4 ± 0.7 | 25.0 ± 1.1 | 24.8 ± 1.0 | 29.8 ± 0.9 | ||
| K119A | 19.5 ± 0.4 | 24.4 ± 0.6 | 24.7 ± 0.2 | 31.38 ± 0.05 | |||
| V122A | 15.8 ± 0.2 | 27.1 ± 1.3 | 30.3 ± 0.6 | 26.7 ± 2.1 | |||
| Protein | T1/2, °C | Protein | T1/2, °C | ||||
|---|---|---|---|---|---|---|---|
| N-terminal domain | ‘black cluster’ | rWT | 64.3 ± 2.5 | C-terminal domain | ‘black cluster’ | ||
| F35A | 59.8 ± 1.2 | F106A | 61.2 ± 0.6 | ||||
| F83A | 56.8 ± 0.1 | E169A | 62.6 ± 3.5 | ||||
| Y86A | 67.8 ± 1.3 | F172A | 53.4 ± 0.2 | ||||
| ‘gray cluster’ | F70A | 66.5 ± 1.4 | ‘gray cluster’ | W156A | 48.6 ± 0.8 | ||
| Q46A | 60.4 ± 0.5 | K119A | 55.1 ± 0.9 | ||||
| F49A | 66.9 ± 1.8 | V122A | 59.0 ± 0.6 | ||||
| Protein | Sequential Binding Model | Cooperative Binding Model | |||
|---|---|---|---|---|---|
| log K1 | log K2 | log K | n | ||
| rWT | 4.5 | 4.1 | 4.6 | 1.3 | |
| N-terminal domain | |||||
| ‘black’ cluster | F35A | 4.6 | 4.8 | 4.7 | 1.5 |
| F83A | 5.8 | 2.8 | 5,8 | 0.9 | |
| Y86A | 6.2 | 4.5 | n/d | n/d | |
| ‘gray’ cluster | F70A | 3.2 | 3.4 | n/a | n/a |
| Q46A | 4.7 | n/a | n/a | ||
| F49A | 4.4 | 3.3 | 4.1 | 0.8 | |
| C-terminal domain | |||||
| ‘black’ cluster | F106A | 3.9 | 4.0 | 3.9 | 1.4 |
| E169A | 4.4 | 4.1 | 4.6 | 1.4 | |
| F172A | n/d | n/d | 4.0 | 1.7 | |
| ‘gray’ cluster | W156A | n/d | n/d | 3.5 | 1.9 |
| K119A | 8.7 | 7.9 | 7.9 | 1.1 | |
| V122A | 4.0 | 4.2 | 4.2 | 1.4 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Permyakov, S.E.; Vologzhannikova, A.S.; Nemashkalova, E.L.; Kazakov, A.S.; Denesyuk, A.I.; Denessiouk, K.; Baksheeva, V.E.; Zamyatnin, A.A., Jr.; Zernii, E.Y.; Uversky, V.N.; et al. Experimental Insight into the Structural and Functional Roles of the ‘Black’ and ‘Gray’ Clusters in Recoverin, a Calcium Binding Protein with Four EF-Hand Motifs. Molecules 2019, 24, 2494. https://doi.org/10.3390/molecules24132494
Permyakov SE, Vologzhannikova AS, Nemashkalova EL, Kazakov AS, Denesyuk AI, Denessiouk K, Baksheeva VE, Zamyatnin AA Jr., Zernii EY, Uversky VN, et al. Experimental Insight into the Structural and Functional Roles of the ‘Black’ and ‘Gray’ Clusters in Recoverin, a Calcium Binding Protein with Four EF-Hand Motifs. Molecules. 2019; 24(13):2494. https://doi.org/10.3390/molecules24132494
Chicago/Turabian StylePermyakov, Sergey E., Alisa S. Vologzhannikova, Ekaterina L. Nemashkalova, Alexei S. Kazakov, Alexander I. Denesyuk, Konstantin Denessiouk, Viktoriia E. Baksheeva, Andrey A. Zamyatnin, Jr., Evgeni Yu. Zernii, Vladimir N. Uversky, and et al. 2019. "Experimental Insight into the Structural and Functional Roles of the ‘Black’ and ‘Gray’ Clusters in Recoverin, a Calcium Binding Protein with Four EF-Hand Motifs" Molecules 24, no. 13: 2494. https://doi.org/10.3390/molecules24132494
APA StylePermyakov, S. E., Vologzhannikova, A. S., Nemashkalova, E. L., Kazakov, A. S., Denesyuk, A. I., Denessiouk, K., Baksheeva, V. E., Zamyatnin, A. A., Jr., Zernii, E. Y., Uversky, V. N., & Permyakov, E. A. (2019). Experimental Insight into the Structural and Functional Roles of the ‘Black’ and ‘Gray’ Clusters in Recoverin, a Calcium Binding Protein with Four EF-Hand Motifs. Molecules, 24(13), 2494. https://doi.org/10.3390/molecules24132494









