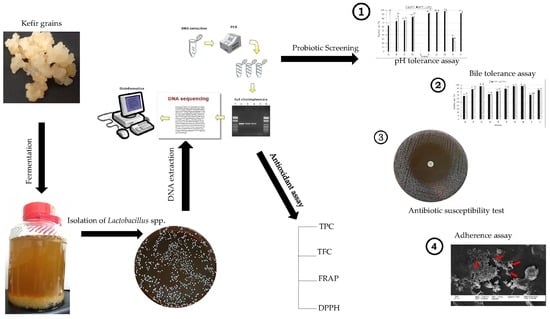
Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia
Abstract
:1. Introduction
2. Results
2.1. Isolation and Identification of Lactobacillus spp. from Kefir
2.2. Tolerance to Low pH Condition of the Isolates
2.3. Bile Salt Tolerance Test
2.4. Adherence Assay
2.5. Scanning Electron Microscopy of Adhered Isolated Lactobacillus spp. toward HT-29 Cells
2.6. Antibiotic Susceptibility Test
2.7. Hemolytic Test
2.8. Antioxidant Assay
3. Discussion
4. Materials and Methods
4.1. Kefir Grains
4.2. Enumeration and Isolation of Lactic Acid Bacteria from Kefir Grains
4.3. Lactic Acid Bacteria Identification using 16S rRNA Sequence Analyses
4.4. Determination Probiotics Properties of Isolated Lactobacillus spp. in the Gastrointestinal Tract Model
4.4.1. Tolerance to Low pH Conditions
4.4.2. Bile Salt Tolerance Test
4.4.3. Adherence Assay
4.4.4. Scanning Electron Microscopy (SEM) of Adhered Isolated Lactobacillus toward Intestinal Cells
4.4.5. Antibiotic Susceptibility Test
4.4.6. Hemolytic Test
4.5. Determination of Antioxidant Activity
4.5.1. Total Phenolic Content (TPC) Assay
4.5.2. Total Flavonoid Content (TFC) Assay
4.5.3. 2,2-Diphenyl-2-picrylhydrazyl Assay
4.5.4. Ferric Reducing Antioxidant Power Assay
4.6. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligne, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Nuraida, L. A review: Health promoting lactic acid bacteria in traditional Indonesian fermented foods. Food Sci. Hum. Wellness 2015, 4, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, B.E.; Dutton, R.J. Fermented foods as experimentally tractable microbial ecosystems. Cell 2015, 161, 49–55. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V. From yaks to yogurt: The history, development, and current use of probiotics. Clin. Infect. Dis. 2015, 60, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Stanton, C.; Ross, R.P.; Fitzgerald, G.F.; Van Sinderen, D. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 2005, 16, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of Action of Probiotics. Adv. Nutr. 2019, 10, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Sen, M. Role of Probiotics in Health and Disease–A Review. Int. J. Adv. Life Sci. Res. 2019, 1–11. [Google Scholar]
- Thakur, K.; Zhang, J.G.; Wei, Z.J.; Kumar, N.; Tomar, S.K.; Pophaly, S.D. CrossTalk Between Functional Foods and Gut Health. In Complementary and Alternative Medicine: Breakthroughs in Research and Practice; IGI Global: Hershey, PA, USA, 2018; pp. 330–351. ISBN 9781522529705. [Google Scholar]
- Tripathi, M.K.; Giri, S.K. Probiotic functional foods: Survival of probiotics during processing and storage. J. Funct. Foods 2014, 9, 225–241. [Google Scholar] [CrossRef]
- Oh, N.S.; Joung, J.Y.; Lee, J.Y.; Kim, Y. Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PLoS ONE 2018, 13, 2. [Google Scholar] [CrossRef]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. Lwt Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Sharifi, M.; Moridnia, A.; Mortazavi, D.; Salehi, M.; Bagheri, M.; Sheikhi, A. Kefir: A powerful probiotics with anticancer properties. Med Oncol. 2017, 34, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yovanoudi, M.; Dimitreli, G.; Raphaelides, S.N.; Antoniou, K.D. Flow behavior studies of kefir type systems. J. Food Eng. 2013, 118, 41–48. [Google Scholar] [CrossRef]
- Gao, X.; Li, B. Chemical and microbiological characteristics of kefir grains and their fermented dairy products: A review. Food Agric. 2017, 1–10. [Google Scholar] [CrossRef]
- Prado, M.R.; Blandón, L.M.; Vandenberghe, L.P.S.; Rodrigues, C.; Castro, G.R.; Thomaz-Soccol, V.; Soccol, C.R. Milk kefir: Composition, microbial cultures, biological activities, and related products. Front. Microbiol. 2015, 6, 1177. [Google Scholar] [CrossRef] [PubMed]
- Bengoa, A.A.; Zavala, L.; Carasi, P.; Trejo, S.A.; Bronsoms, S.; Serradell, M.L.Á.; Garrote, G.L.; Abraham, A.G. Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res. Int. 2018, 103, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Jeong, D.; Kang, I.B.; Kim, H.; Song, K.Y.; Seo, K.H. Dual function of Lactobacillus kefiri DH5 in preventing high-fat-diet-induced obesity: Direct reduction of cholesterol and upregulation of PPAR-α in adipose tissue. Mol. Nutr. Food Res. 2017, 61, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, I.; Dolar, M.E.; Ozpinar, H. Effect of administering kefir on the changes in fecal microbiota and symptoms of inflammatory bowel disease: A randomized controlled trial. Turk. J. Gastroenterol. 2018, 30, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, A.; Tsakiris, A.; Kanellaki, M.; Bezirtzoglou, E.; Akrida-Demertzi, K.; Kourkoutas, Y. Wine production using free and immobilized kefir culture on natural supports. Food Chem. 2019, 272, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Yépez, A.; Russo, P.; Spano, G.; Khomenko, I.; Biasioli, F.; Capozzi, V.; Aznar, R. In situ riboflavin fortification of different kefir-like cereal-based beverages using selected Andean LAB strains. Food Microbiol. 2019, 77, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Jaya, F.; Thohari, I.; Susilorini, T.E.; Asmara, D.R. Microbiological properties of preparing facial mask cream from goat milk kefir. Iop Conf. Ser. Earth Environ. Sci. 2019, 230, 012105. [Google Scholar] [CrossRef]
- Erdoğdular, A.O.; Apar, D.K. Bioremoval of reactive dye Remazol Navy by kefir grains. Appl. Biol. Chemistry. 2019, 62, 1–11. [Google Scholar] [CrossRef]
- Zanirati, D.F.; Abatemarco, M.; de Cicco Sandes, S.H.; Nicoli, J.R.; Nunes, Á.C.; Neumann, E. Selection of lactic acid bacteria from Brazilian kefir grains for potential use as starter or probiotic cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, I.; Chondrou, P.; Bontsidis, C.; Karolidou, K.; Terpou, A.; Alexopoulos, A.; Bezirtzoglou, E.; Galanis, A.; Plessas, S. Assessment of the probiotic potential of lactic acid bacteria isolated from kefir grains: Evaluation of adhesion and anti-proliferative properties in in vitro experimental systems. Ann. Microbiol. 2019, 1–13. [Google Scholar] [CrossRef]
- Yerlikaya, O. Probiotic potential and biochemical and technological properties of Lactococcus lactis ssp. lactis strains isolated from raw milk and kefir grains. J. Dairy Sci. 2018, 102, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Pringsulaka, O.; Rueangyotchanthana, K.; Suwannasai, N. In vitro screening of lactic acid bacteria for multi-strain probiotics. Livest. Sci. 2015, 174, 66–73. [Google Scholar] [CrossRef]
- Arslan, S. A review: Chemical, microbiological and nutritional characteristics of kefir. Cyta - J. Food 2015, 13, 340–345. [Google Scholar] [CrossRef]
- Garofalo, C.; Osimani, A.; Milanovi, V.; Aquilanti, L.; De Filippis, F.; Stellato, G.; Di Mauro, S.; Turchetti, B.; Buzzini, P.; Ercolini, D.; et al. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 2015, 49, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Sahadeva, R.P.K.; Leong, S.F.; Chua, K.H.; Tan, C.H.; Chan, H.Y.; Tong, E.V.; Wong, S.Y.W.; Chan, H.K. Survival of commercial probiotic strains to pH and bile. Int. Food Res. J. 2011, 18, 1515–1522. [Google Scholar]
- Schillinger, U.; Guigas, C.; Holzapfel, W.H. In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 2005, 15, 1289–1297. [Google Scholar] [CrossRef]
- Huang, Y.; Adams, M.C. In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int. J. Food Microbiol. 2004, 91, 253–260. [Google Scholar] [CrossRef]
- Lin, W.; Hwang, C.; Chen, L.; Tsen, H. Viable counts, characteristic evaluation for commercial lactic acid bacteria products. Food Microbiol. 2006, 23, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Ilha, E.C.; da Silva, T.; Lorenz, J.G.; de Oliveira Rocha, G.; Sant’Anna, E.S. Lactobacillus paracasei isolated from grape sourdough: Acid, bile, salt, and heat tolerance after spray drying with skim milk and cheese whey. Eur. Food Res. Technol. 2006, 240, 977–984. [Google Scholar] [CrossRef]
- Gao, Y.; Li, D.; Liu, S.; Liu, Y. Probiotic potential of L. sake C2 isolated from traditional Chinese fermented cabbage. Eur. Food Res. Technol. 2012, 234, 45–51. [Google Scholar] [CrossRef]
- Prasad, J.; Gill, H.; Smart, J.; Gopal, P.K. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int. Dairy J. 1999, 8, 993–1002. [Google Scholar] [CrossRef]
- Tokatlı, M.; Gülgör, G.; Bağder Elmacı, S.; Arslankoz İşleyen, N.; Özçelik, F. In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. Biomed Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zielińska, D.; Rzepkowska, A.; Radawska, A.; Zieliński, K. In vitro screening of selected probiotic properties of Lactobacillus strains isolated from traditional fermented cabbage and cucumber. Curr. Microbiol. 2015, 70, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, A.S.N.; Jagadeesh, K.S.; Krishnaraj, P.U. Isolation and Screening of Lactic Acid Bacteria for Acidic pH and Bile Tolerance. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3975–3980. [Google Scholar] [CrossRef]
- Roda, E.; Aldini, R.; Mazzella, G.; Roda, A.; Sama, C.; Festi, D.; Barbara, L. Enterohepatic circulation of bile acids after cholecystectomy. Gut 1978, 19, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Archer, A.C.; Halami, P.M. Probiotic attributes of Lactobacillus fermentum isolated from human feces and dairy products. Appl. Microbiol. Biotechnol. 2015, 99, 8113–8123. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, I.; Moussa, O.B.; Khaldi, T.E.; Kebouchi, M.; Soligot-Hognon, C.; Leroux, Y.; Hassouna, M. Adhesion Properties of Probiotic Lactobacillus Strains Isolated from Tunisian Sheep and Goat Milk. J. Agric. Sci. Technol. 2019, 21, 587–600. [Google Scholar]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Jahromi, M.F.; Ho, Y.W. Probiotic Potential of Lactobacillus Strains with Antimicrobial Activity against Some Human Pathogenic Strains. Biomed Res. Int. 2014. [Google Scholar]
- Archer, A.C.; Kurrey, N.K.; Halami, P.M. In vitro adhesion and anti-inflammatory properties of native Lactobacillus fermentum and Lactobacillus delbrueckiispp. J. Appl. Microbiol. 2018, 125, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Sánchez, B.; Suárez, J.E.; Urdaci, M.C. Characterization of the adherence properties of human Lactobacilli strains to be used as vaginal probiotics. Fems Microbiol. Lett. 2012, 328, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Muryany, I.; Lian, H.H.; Ina-Salwany, A.R.G.; Ghazali, A.R.; Zamri-Saad, M.; Rajab, N.F. Adhesion Ability and Cytotoxic Evaluation of Lactobacillus Strains Isolated from Malaysian Fermented Fish (Pekasam) on Ht-29 and Ccd-18Co Intestinal Cells. Sains Malays. 2018, 47, 2391–2399. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, R.; Yocheva, L.; Tserovska, L.; Zhelezova, G.; Stefanova, N.; Atanasova, A.; Danguleva, A.; Ivanova, G.; Karapetkov, N.; Rumyan, N.; et al. Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol. Biotechnol. Equip. 2015, 29, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Padmavathi, T.; Bhargavi, R.; Priyanka, P.R.; Niranjan, N.R.; Pavitra, P.V. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J. Genet. Eng. Biotechnol. 2018, 16, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; Bai, Y.; Di Cagno, R.; Gobbetti, M.; Gänzle, M.G. Metabolism of phenolic compounds by Lactobacillus spp. during fermentation of cherry juice and broccoli puree. Food Microbiol. 2015, 46, 272–279. [Google Scholar] [CrossRef]
- Bhanja Dey, T.; Chakraborty, S.; Jain, K.K.; Sharma, A.; Kuhad, R.C. Antioxidant phenolics and their microbial production by submerged and solid state fermentation process: A review. Trends Food Sci. Technol. 2016, 53, 60–74. [Google Scholar] [CrossRef]
- Muñoz, R.; de las Rivas, B.; López de Felipe, F.; Reverón, I.; Santamaría, L.; Esteban-Torres, M.; Curiel, J.A.; Rodríguez, H.; Landete, J.M. Biotransformation of Phenolics by Lactobacillus plantarum in Fermented Foods. Fermented Foods Health Dis. Prev. 2016, 63–83. [Google Scholar]
- Zulkawi, N.; Ng, K.H.; Zamberi, R.; Yeap, S.K.; Satharasinghe, D.; Jaganath, I.B.; Jamaluddin, A.B.; Tan, S.W.; Ho, W.Y.; Alitheen, N.B.; et al. In vitro characterization and in vivo toxicity, antioxidant and immunomodulatory effect of fermented foods; Xeniji™. Bmc Complementary Altern. Med. 2017, 17, 344. [Google Scholar] [CrossRef] [PubMed]
- Adetuyi, F.O.; Ibrahim, T.A. Effect of Fermentation Time on the Phenolic, Flavonoid and Vitamin C Contents and Antioxidant Activities of Okra (Abelmoschus esculentus) Seeds. Niger. Food J. 2015, 32, 128–137. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, L.; Rui, X.; Li, W.; Chen, X.; Jiang, M.; Dong, M. Enhancement of the antioxidant capacity of soy whey by fermentation with Lactobacillus plantarum B1–6. J. Funct. Foods 2015, 12, 33–44. [Google Scholar] [CrossRef]
- Thaipong, K.; Boonprakob, U.; Crosby, K.; Cisneros-Zevallos, L.; Hawkins Byrne, D. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compos. Anal. 2006, 19, 669–675. [Google Scholar] [CrossRef]
- Oh, N.S.; Lee, J.Y.; Oh, S.; Joung, J.Y.; Kim, S.G.; Shin, Y.K.; Lee, K.W.; Kim, S.H.; Kim, Y. Improved functionality of fermented milk is mediated by the synbiotic interaction between Cudrania tricuspidata leaf extract and Lactobacillus gasseri strains. Appl. Microbiol. Biotechnol. 2016, 100, 5919–5932. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, T.; Pihlanto, A.; Akkanen, S.; Korhonen, H. Development of antioxidant activity in milk whey during fermentation with lactic acid bacteria. J. Appl. Microbiol. 2007, 102, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cai, Y.; Zhou, G.; Shi, X.; Su, J.; Chen, G.; Lin, K. Rapid Sanger Sequencing of the 16S rRNA Gene for Identification of Some Common Pathogens. PLoS ONE 2014, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Leite, A.M.O.; Miguel, M.A.L.; Peixoto, R.S.; Paschoalin, V.M.F.; Mayo, B. Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J. Dairy Sci. 2015, 98, 3622–3632. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Baik, S. Probiotic properties of Lactobacillus strains with high cinnamoyl esterase activity isolated from jeot-gal, a high-salt fermented seafood. Ann. Microbiol. 2019, 1–11. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Hayat, H.F.; Sarwar, S.; Mehwish, H.M.; Ahmad, F.; Hussain, N.; Shah, S.Z.H.; Khurshid, M.; Siddiqu, M.; Shi, J. Isolation and evaluation of probiotic potential of lactic acid bacteria isolated from poultry intestine. Microbiology 2018, 87, 116–126. [Google Scholar] [CrossRef]
- Ho, W.Y.; Beh, B.K.; Lim, K.L.; Mohamad, N.E.; Yusof, H.M.; Ky, H.; Tan, S.W.; Jamaluddin, A.; Long, K.; Lim, C.L.; et al. Antioxidant and hepatoprotective effects of the food seasoning curry leaves Murraya Koenigii (L.) Spreng (Rutaceae). RSC Adv. 2015, 5, 100589–100597. [Google Scholar] [CrossRef]
Sample Availability: Samples of kefir are available from the authors. |









| 16S rRNA Sequences from Isolates | Accession No. | Matches to 16S rRNA Sequences from GenBank Database | Identity (%) with GenBank Database |
|---|---|---|---|
| A | KC415613.1 | L. harbinensis strain B22 | 99% |
| KX279364.1 | L. harbinensis strain HBUAS5305 | 99% | |
| MH393129.1 | L. harbinensis strain NBRC 100982 | 99% | |
| B | KC415613.1 | L. harbinensis strain B22 | 99% |
| AY974809.1 | L. brevis strain HDRS2 | 99% | |
| KF418816.1 | L. harbinensis strain FQ003 | 99% | |
| C | KC415613.1 | L. harbinensis strain B22 | 99% |
| KX279364.1 | L. harbinensis strain HBUAS5305 | 99% | |
| KX279985.1 | L. sp MS6 | 98% | |
| D | EU559596.1 | L. plantarum Gt2 | 98% |
| KY041688.1 | L. plantarum ZDY36a | 98% | |
| MH472974.1 | L. plantarum HBUAS52249 | 98% | |
| E | MH392958.1 | L. paracasei strain HBUAS52231 | 98% |
| MH472956.1 | L. paracasei strain HBUAS53273 | 98% | |
| MF083138.1 | L. casei strain YQ116 | 98% | |
| F | KC415613.1 | L. harbinensis strain B22 | 99% |
| KX279985.1 | L. sp MS6 | 99% | |
| AY974809.1 | L. brevis strain HDRS2 | 99% | |
| G | MH472956.1 | L. paracasei strain HBUAS52231 | 98% |
| MH392958.1 | L. paracasei strain HBUAS52231 | 98% | |
| KU851192.1 | L. casei strain H19.9 | 98% | |
| H | MG551235.1 | L. plantarum strain NWAFU1558 | 98% |
| KJ736728.1 | L. plantarum strain Akhavan-Q3 | 98% | |
| EU637397.1 | L. plantarum strain Y-2-9 | 97% | |
| I | MH392958.1 | L. paracasei strain HBUAS52231 | 99% |
| MH472956.1 | L. paracasei strain HBUAS53273 | 99% | |
| MF083138.1 | L. casei strain YQ116 | 98% | |
| J | MH620395.1 | L. plantarum strain MSD1-4 | 98% |
| CP0222294.1 | L. plantarum strain DSR M2 | 98% | |
| CP028977.1 | L. plantarum strain LQ80 | 98% |
| Isolate | Diameter Zone Inhibition (mm) | ||||
|---|---|---|---|---|---|
| VA | CN | AMP | TE | P | |
| A | - | - | +++ | +++ | +++ |
| B | - | - | +++ | +++ | +++ |
| C | - | - | +++ | +++ | +++ |
| D | - | - | +++ | +++ | +++ |
| E | - | - | +++ | +++ | +++ |
| F | - | - | +++ | +++ | +++ |
| G | - | - | +++ | +++ | +++ |
| H | - | - | +++ | +++ | +++ |
| I | - | - | +++ | +++ | +++ |
| J | - | - | +++ | +++ | +++ |
| Isolate | TPC (mg GAE/mg protein) | TFC (mg catechin/mg protein) | FRAP (mM FRAP/μg protein) | DPPH (%) |
|---|---|---|---|---|
| A | 21.94 ± 4.52 a | 9.12 ± 0.46 a,b | 0.25 ± 0.01 a | 63.10 ± 0.20 e |
| B | 58.26 ± 4.42 c,d | 10.39 ± 0.28 b,c | 0.97 ± 0.01 c | 54.33 ± 0.36 b |
| C | 45.41 ± 1.36 b,c | 10.76 ± 0.99 b,c | 1.46 ± 0.14 d | 60.21 ± 0.11 d |
| D | 39.71 ± 3.10 b | 7.51 ± 0.35 a | 0.64 ± 0.14 b | 48.43 ± 0.73 a |
| E | 60.42 ± 2.69 d | 12.61 ± 0.46 c | 1.25 ± 0.01 d | 53.67 ± 0.20 b |
| F | 75.14 ± 8.58 e | 19.13 ± 0.04 d | 1.76 ± 0.10 e | 67.40 ± 0.81 f |
| G | 115.97 ± 7.22 g,* | 58.94 ± 2.06 f,* | 2.81 ± 0.07 f | 76.79 ± 0.47 g,* |
| H | 99.20 ± 4.46 f | 36.29 ± 0.82 e | 2.58 ± 0.08 f | 58.70 ± 0.63 c |
| I | 50.93 ± 4.65 b,c,d | 12.24 ± 0.43 c | 1.81 ± 0.02 e | 48.91 ± 0.24 a |
| J | 39.93 ± 1.73 b | 7.72 ± 0.37 a | 0.79 ± 0.005 b,c | 49.50 ± 0.19 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talib, N.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia. Molecules 2019, 24, 2606. https://doi.org/10.3390/molecules24142606
Talib N, Mohamad NE, Yeap SK, Hussin Y, Aziz MNM, Masarudin MJ, Sharifuddin SA, Hui YW, Ho CL, Alitheen NB. Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia. Molecules. 2019; 24(14):2606. https://doi.org/10.3390/molecules24142606
Chicago/Turabian StyleTalib, Noorshafadzilah, Nurul Elyani Mohamad, Swee Keong Yeap, Yazmin Hussin, Muhammad Nazirul Mubin Aziz, Mas Jaffri Masarudin, Shaiful Adzni Sharifuddin, Yew Woh Hui, Chai Ling Ho, and Noorjahan Banu Alitheen. 2019. "Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia" Molecules 24, no. 14: 2606. https://doi.org/10.3390/molecules24142606
APA StyleTalib, N., Mohamad, N. E., Yeap, S. K., Hussin, Y., Aziz, M. N. M., Masarudin, M. J., Sharifuddin, S. A., Hui, Y. W., Ho, C. L., & Alitheen, N. B. (2019). Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia. Molecules, 24(14), 2606. https://doi.org/10.3390/molecules24142606