Discovery of Novel DPP-IV Inhibitors as Potential Candidates for the Treatment of Type 2 Diabetes Mellitus Predicted by 3D QSAR Pharmacophore Models, Molecular Docking and De Novo Evolution
Abstract
1. Introduction
2. Results and Discussion
2.1. Generation of Pharmacophore Models
2.2. Validation of Pharmacophore Models
2.2.1. Training and Test Sets
2.2.2. Fischer’s Randomization Test
2.2.3. Virtual Screening
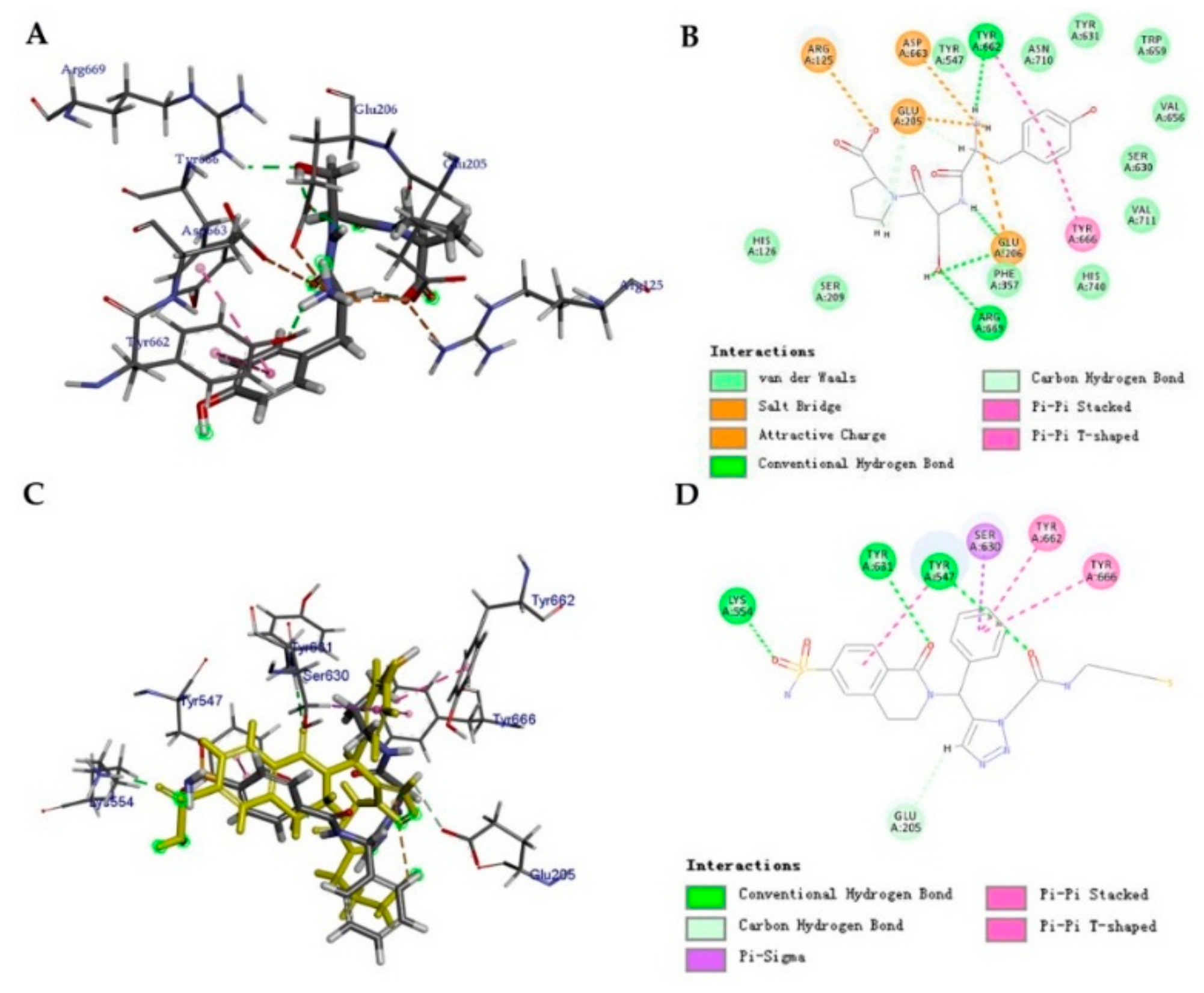
2.3. Molecular Docking Analysis
2.4. Ligand de novo Evolution Analysis
2.5. Prediction activity (IC50) of Compounds from Databases and their Derivatives Generated by de novo Evolution.
2.6. Calculation of RMSD of Re-Docking and Cross-Docking Tests
3. Materials and Methods
3.1. Data Preparation
3.2. Pharmacophore Modeling
3.3. Pharmacophore Model Evaluation
3.3.1. Cost Analysis
3.3.2. Fischer Validation
3.4. Virtual Screening
3.5. Docking Protocol
3.6. De Novo Evolution Protocol
3.7. Re-docking & Cross-docking Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Meduru, H.; Wang, Y.-T.; Tsai, J.J.P.; Chen, Y.-C. Finding a Potential Dipeptidyl Peptidase-4 (DPP-4) Inhibitor for Type-2 Diabetes Treatment Based on Molecular Docking, Pharmacophore Generation, and Molecular Dynamics Simulation. Inter. J. Molecular Sci. 2016, 17, 920. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation-What is Diabetes. Available online: https://www.idf.org/about-diabetes/what-is-diabetes.html (accessed on 27 April 2018).
- Amini, Z.; Fatemi, M.H.; Gharaghani, S. Hybrid docking-QSAR studies of DPP-IV inhibition activities of a series of aminomethyl-piperidones. Comput. Biol. Chem. 2016, 64, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Pathak, R.; Bridgeman, M.B. Dipeptidyl Peptidase-4 (DPP-4) Inhibitors In the Management of Diabetes. Pharm.Ther. 2010, 35, 509–513. [Google Scholar] [PubMed]
- Andersen, E.S.; Deacon, C.F.; Holst, J.J. Do we know the true mechanism of action of the DPP-4 inhibitors? Diabetes Obes. Metab. 2018, 20, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-R.; Kim, H.Y.; Choi, I.; Kim, J.-B.; Jin, C.H.; Han, A.-R. DPP-IV Inhibitory Potentials of Flavonol Glycosides Isolated from the Seeds of Lens culinaris: In Vitro and Molecular Docking Analyses. Molecules 2018, 23, 1998. [Google Scholar] [CrossRef]
- Li, G.; Huan, Y.; Yuan, B.; Wang, J.; Jiang, Q.; Lin, Z.; Shen, Z.; Huang, H. Discovery of novel xanthine compounds targeting DPP-IV and GPR119 as anti-diabetic agents. Eur. J. Med. Chem. 2016, 124, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Schnapp, G.; Klein, T.; Hoevels, Y.; Bakker, R.A.; Nar, H. Comparative Analysis of Binding Kinetics and Thermodynamics of Dipeptidyl Peptidase-4 Inhibitors and Their Relationship to Structure. J. Med. Chem. 2016, 59, 7466–7477. [Google Scholar] [CrossRef]
- Uchida, T.; Wakasugi, M.; Kitamura, T.; Yamamoto, T.; Asakura, M.; Fujiwara, R.; Itoh, T.; Fujii, H.; Hirono, S. Exploration of DPP-IV inhibitors with a novel scaffold by multistep in silico screening. J. Mol. Graphics Model. 2018, 79, 254–263. [Google Scholar] [CrossRef]
- Dube, D.; Periwal, V.; Kumar, M.; Sharma, S.; Singh, T.P.; Kaur, P. 3D-QSAR based pharmacophore modeling and virtual screening for identification of novel pteridine reductase inhibitors. J. Mol. Model. 2012, 18, 1701–1711. [Google Scholar] [CrossRef]
- Al-masri, I.M.; Mohammad, K.M.; Taha, M.O. Discovery of DPP IV Inhibitors by Pharmacophore Modeling and QSAR Analysis followed by in silico Screening. Chem. Med. Chem. 2008, 3, 1763–1779. [Google Scholar] [CrossRef]
- Ataei, S.; Yilmaz, S.; Ertan-Bolelli, T.; Yildiz, I. Generated 3D-common feature hypotheses using the HipHop method for developing new topoisomerase I inhibitors. Arch. Pharm. (Weinheim) 2015, 348, 498–507. [Google Scholar] [CrossRef] [PubMed]
- TMC Library Traditional Chinese Medicine Database (TCM). Available online: https://library.tmc.edu/database/traditional-chinese-medicine-database-tcm/ (accessed on 7 July 2019).
- Desaphy, J.; Bret, G.; Rognan, D.; Kellenberger, E. sc-PDB: A 3D-database of ligandable binding sites—10 years on. Nucleic Acids Res. 2015, 43, D399–D404. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.M.; Chu, H.D.; Kuethe, J.T.; Gao, Y.-D.; Scapin, G.; Eiermann, G.; He, H.; Li, X.; Lyons, K.A.; Metzger, J.; et al. The discovery of novel 5,6,5-and 5,5,6-tricyclic pyrrolidines as potent and selective DPP-4 inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 2622–2626. [Google Scholar] [CrossRef] [PubMed]
- Jadav, P.; Bahekar, R.; Shah, S.R.; Patel, D.; Joharapurkar, A.; Jain, M.; Sairam, K.V.V.M.; Singh, P.K. Design, synthesis and biological evaluation of novel aminomethyl-piperidones based DPP-IV inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 1918–1922. [Google Scholar] [CrossRef]
- Jiang, C.; Han, S.; Chen, T.; Chen, J. 3D-QSAR and docking studies of arylmethylamine-based DPP IV inhibitors. Acta Pharmaceutica Sinica B 2012, 2, 411–420. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Han, L.; Zhang, B.; Zhou, J.; Zhang, H. Synthesis and biological evaluation of triazole based uracil derivatives as novel DPP-4 inhibitors. Org. Biomol. Chem. 2016, 14, 9598–9611. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, H.; Cui, S.; Wu, F.; Zhang, Y.; Su, M.; Gong, Y.; Qiu, S.; Jiao, Q.; Qin, C.; et al. Discovery and Rational Design of Natural-Product-Derived 2-Phenyl-3,4-dihydro-2H-benzo[f]chromen-3-amine Analogs as Novel and Potent Dipeptidyl Peptidase 4 (DPP-4) Inhibitors for the Treatment of Type 2 Diabetes. J. Med. Chem. 2016, 59, 6772–6790. [Google Scholar] [CrossRef] [PubMed]
- Sawant, S.D.; Nerkar, A.; Velapure, A.V.; Pawar, N.D. Design, synthesis, QSAR studies and in vitro evaluation of novel triazolopiperazine based B-Amino amides as dipeptidyl peptidase-IV (DPP-IV) inhibitors: Part-I. Int. J. Pharm.Pharm. Sci. 2014, 6, 760–765. [Google Scholar]
- Sutton, J.M.; Clark, D.E.; Dunsdon, S.J.; Fenton, G.; Fillmore, A.; Harris, N.V.; Higgs, C.; Hurley, C.A.; Krintel, S.L.; MacKenzie, R.E.; et al. Novel heterocyclic DPP-4 inhibitors for the treatment of type 2 diabetes (vol 22, pg 1464, 2012). Bioorg. Med. Chem. Lett. 2012, 22, 2359. [Google Scholar] [CrossRef]
- Zhang, Z.; Wallace, M.B.; Feng, J.; Stafford, J.A.; Skene, R.J.; Shi, L.; Lee, B.; Aertgeerts, K.; Jennings, A.; Xu, R.; et al. Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV. J. Med. Chem. 2011, 54, 510–524. [Google Scholar] [CrossRef] [PubMed]
- Scior, T.; Medina-Franco, J.L.; Do, Q.-T.; Martínez-Mayorga, K.; Yunes Rojas, J.A.; Bernard, P. How to recognize and workaround pitfalls in QSAR studies: A critical review. Curr. Med. Chem. 2009, 16, 4297–4313. [Google Scholar] [CrossRef] [PubMed]
- Rampogu, S.; Lee, G.; Baek, A.; Son, M.; Park, C.; Zeb, A.; Yoon, S.H.; Park, S.; Lee, K.W. Discovery of Non-Peptidic Compounds against Chagas Disease Applying Pharmacophore Guided Molecular Modelling Approaches. Molecules 2018, 23, 3054. [Google Scholar] [CrossRef] [PubMed]
- Ashry, E.S.H.E.; Badawy, M.E.I.; El-kilany, Y.; Nahas, N.M.; Al-Ghamdi, M.A. 3D-QSAR Pharmacophore-based Ligand Alignment, Virtual Screening and Molecular Docking of Arylidene (Benzimidazol-1-yl)acetohydrazones as Biomimetics of Bacterial Inhibitors. Der. Chem. Sin. 2017, 8. [Google Scholar]
- Nagarajan, S.; Ahmed, A.; Choo, H.; Cho, Y.S.; Oh, K.-S.; Lee, B.H.; Shin, K.J.; Pae, A.N. 3D QSAR pharmacophore model based on diverse IKKβ inhibitors. J. Mol. Model. 2011, 17, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Paliwal, S.K.; Sharma, M.; Mittal, A.; Sharma, S.; Sharma,, J.P. Ligand Based Pharmacophore Modeling, Virtual Screening and Molecular Docking for Identification of Novel CYP51 Inhibitors. Chem. Inform. 2016, 1, 1–10. [Google Scholar]
- Yu, H.; Wang, Z.; Zhang, L.; Zhang, J.; Huang, Q. The discovery of novel vascular endothelial growth factor receptor tyrosine kinases inhibitors: Pharmacophore modeling, virtual screening and docking studies. Chem. Biol. Drug Des. 2007, 69, 204–211. [Google Scholar] [CrossRef]
- Wu, G.S.; Robertson, D.H.; Brooks, C.L.; Vieth, M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER - A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003, 24, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.-Q.; Wang, H.-Y.; Zhao, Y.-L.; Xiang, M.-L.; Jiang, P.-D.; Cao, Z.-X.; Zheng, Y.-Z.; Luo, S.-D.; Yu, L.-T.; Wei, Y.-Q.; et al. Pharmacophore modelling and virtual screening for identification of new aurora-a kinase inhibitors. Chem. Biol. Drug Des. 2008, 71, 533–539. [Google Scholar] [CrossRef]
- Fu, Y.; Sun, Y.-N.; Yi, K.-H.; Li, M.-Q.; Cao, H.-F.; Li, J.-Z.; Ye, F. 3D Pharmacophore-Based Virtual Screening and Docking Approaches toward the Discovery of Novel HPPD Inhibitors. Molecules 2017, 22, 959. [Google Scholar] [CrossRef]
- Sneha, P.; Doss, C.G.P. Gliptins in managing diabetes - Reviewing computational strategy. Life Sci. 2016, 166, 108–120. [Google Scholar] [CrossRef]
- Koska, J.; Spassov, V.Z.; Maynard, A.J.; Yan, L.; Austin, N.; Flook, P.K.; Venkatachalam, C.M. Fully Automated Molecular Mechanics Based Induced Fit Protein−Ligand Docking Method. J. Chem. Inf. Model. 2008, 48, 1965–1973. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

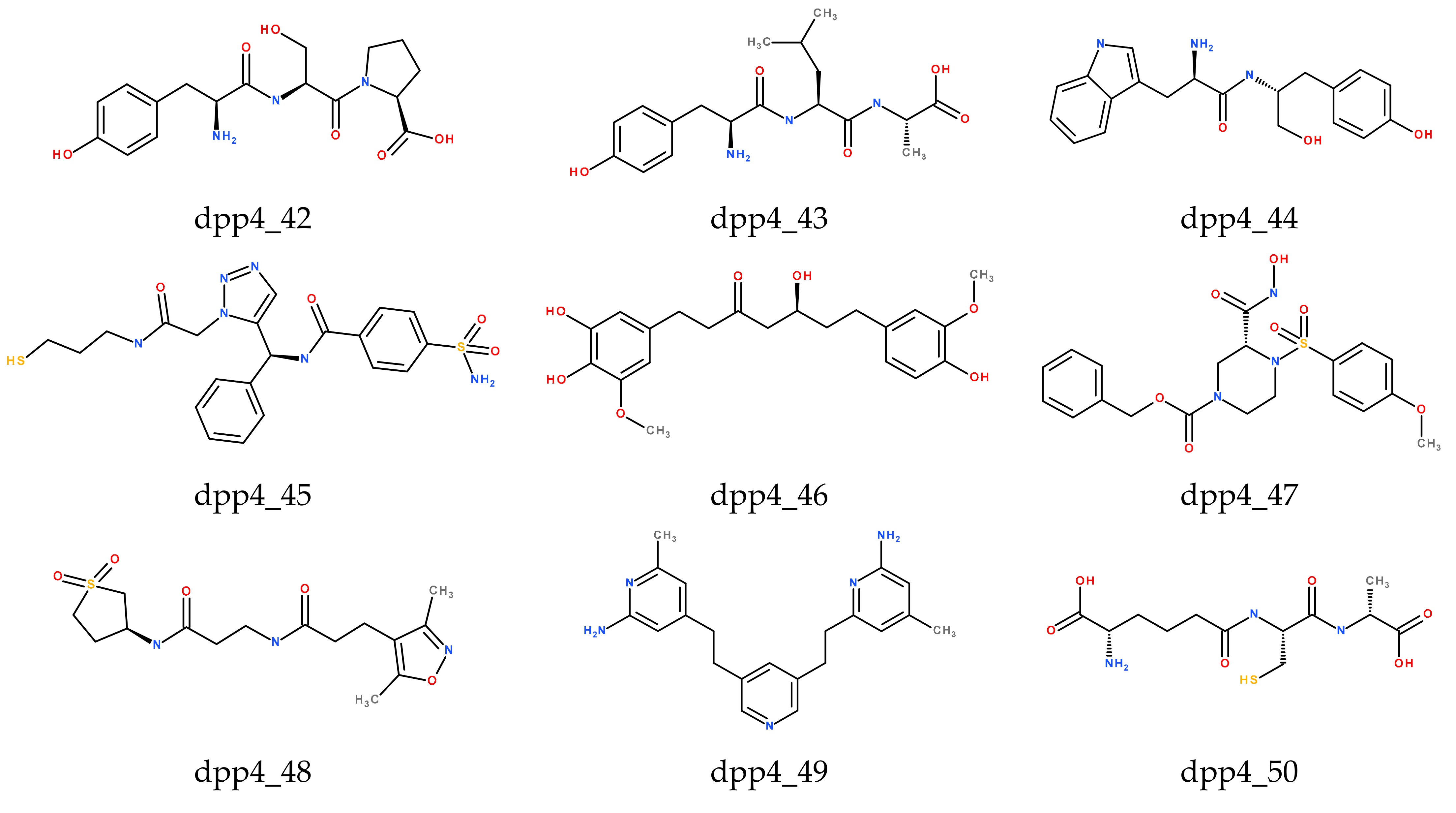


| Compound No. | Fit Value b | Experimental IC50 nM | Predicted IC50 nM | Error a | Experimental Scale c | Predicted Scale c |
|---|---|---|---|---|---|---|
| dpp4_1 | 7.91 | 0.12 | 0.12 | 0 | +++ | +++ |
| dpp4_2 | 7.19 | 0.24 | 0.64 | 0.4 | +++ | +++ |
| dpp4_3 | 6.52 | 2 | 2.99 | 0.99 | +++ | +++ |
| dpp4_4 | 5.88 | 2 | 13 | 11 | +++ | ++ |
| dpp4_5 | 6.13 | 5.8 | 7.31 | 1.51 | +++ | +++ |
| dpp4_6 | 5.98 | 9.56 | 10.52 | 0.96 | +++ | ++ |
| dpp4_7 | 6.09 | 12 | 8.15 | –3.85 | ++ | +++ |
| dpp4_8 | 5.36 | 16 | 43.83 | 27.83 | ++ | ++ |
| dpp4_9 | 5.88 | 17 | 13.16 | –3.84 | ++ | ++ |
| dpp4_10 | 5.71 | 43 | 19.47 | –23.53 | ++ | ++ |
| dpp4_11 | 6.05 | 44 | 8.94 | –35.06 | ++ | +++ |
| dpp4_12 | 4.99 | 45 | 102.83 | 57.83 | ++ | + |
| dpp4_13 | 5.23 | 49.73 | 59.08 | 9.35 | ++ | ++ |
| dpp4_14 | 4.72 | 64.31 | 190.61 | 126.3 | ++ | + |
| dpp4_15 | 5.60 | 120 | 24.86 | –95.14 | + | ++ |
| dpp4_16 | 5.49 | 125 | 32.05 | –92.95 | + | ++ |
| dpp4_17 | 4.88 | 135.45 | 131.69 | –3.76 | + | + |
| dpp4_18 | 4.19 | 180 | 639 | 459 | + | + |
| dpp4_19 | 4.19 | 216.2 | 649.93 | 433.73 | + | + |
| dpp4_20 | 4.83 | 243.67 | 146.8 | –96.87 | + | + |
| dpp4_21 | 4.54 | 443 | 285.39 | –157.61 | + | + |
| dpp4_22 | 4.41 | 540 | 383.37 | –156.63 | + | + |
| dpp4_23 | 4.19 | 800 | 644.7 | –155.3 | + | + |
| dpp4_24 | 4.13 | 1,000 | 736.38 | –263.62 | + | + |
| dpp4_25 | 4.53 | 1,023 | 292.28 | –730.72 | + | + |
| Compound No. | Fit Value b | Experimental IC50 nM | Predicted IC50 nM | Error a | Experimental Scale c | Predicted Scale c |
|---|---|---|---|---|---|---|
| dpp4_26 | 5.88 | 0.6 | 13.15 | 12.55 | +++ | ++ |
| dpp4_27 | 5.93 | 7 | 11.73 | 4.73 | +++ | ++ |
| dpp4_28 | 6.10 | 7.3 | 7.83 | 0.53 | +++ | +++ |
| dpp4_29 | 6.10 | 8.91 | 7.83 | –1.08 | +++ | +++ |
| dpp4_30 | 5.46 | 12.45 | 34.24 | 21.79 | ++ | ++ |
| dpp4_31 | 5.91 | 18 | 12.3 | –5.7 | ++ | ++ |
| dpp4_32 | 5.64 | 23 | 22.72 | –0.28 | ++ | ++ |
| dpp4_33 | 5.94 | 26 | 11.36 | –14.64 | ++ | ++ |
| dpp4_34 | 4.99 | 74 | 100.24 | 26.24 | ++ | + |
| dpp4_35 | 5.11 | 84.72 | 76.97 | –7.75 | ++ | ++ |
| dpp4_36 | 4.85 | 140 | 138.08 | –1.92 | + | + |
| dpp4_37 | 4.55 | 168.63 | 284.03 | 115.4 | + | + |
| dpp4_38 | 4.17 | 340 | 667.67 | 327.67 | + | + |
| dpp4_39 | 4.62 | 452 | 241.7 | –210.3 | + | + |
| dpp4_40 | 4.18 | 990 | 656.11 | -333.89 | + | + |
| dpp4_41 | 4.14 | 1400 | 726.17 | -673.83 | + | + |
| Rank | Name | -CDOCKER | LScore1 | LScore2 | -PLP1 | -PLP2 | -PMF | Consensus |
|---|---|---|---|---|---|---|---|---|
| 1 | dpp4_42 | 61.722 | 6.35 | 6.47 | 104.21 | 103.65 | 157.39 | 6 |
| 2 | dpp4_43 | 83.481 | 5.8 | 6.12 | 87.53 | 79.31 | 136.24 | 4 |
| 3 | dpp4_44 | 47.758 | 5.1 | 5.8 | 94.09 | 93.91 | 132.72 | 1 |
| 4 | dpp4_45 | 46.524 | 4.53 | 5.86 | 103.18 | 86.71 | 119.97 | 1 |
| 5 | dpp4_46 | 42.219 | 5.61 | 6.07 | 79.71 | 82.24 | 116.49 | 1 |
| 6 | dpp4_47 | 40.196 | 4.14 | 5.61 | 87.6 | 80.08 | 128.71 | 0 |
| 7 | dpp4_48 | 46.9 | 3.47 | 5.33 | 74.36 | 66.64 | 130.21 | 0 |
| 8 | dpp4_49 | 43.021 | 4.23 | 5.51 | 75.61 | 69.58 | 109.63 | 0 |
| 9 | dpp4_50 | 74.786 | 5.2 | 5.21 | 62.77 | 52.25 | 95.86 | 1 |
| 10 | dpp4_51 | 42.476 | 3.53 | 5.74 | 69.71 | 57.31 | 119.7 | 0 |
| 11 | Alogliptin * | 40.601 | 3.82 | 6.14 | 96.06 | 92.2 | 142.21 | 4 |
| Rank | Name | -CDOCKER | LScore1 | LScore2 | -PLP1 | -PLP2 | -PMF | Consensus | LUDI3 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | dpp4_42_Evo_1 | 66.80 | 6.62 | 6.82 | 90.23 | 88.58 | 156.25 | 6 | 783 |
| 2 | dpp4_43_Evo_2 | 76.96 | 6.43 | 6.87 | 98.72 | 90.61 | 152.23 | 6 | 508 |
| 3 | dpp4_44_Evo_4 | 26.46 | 4.58 | 6.02 | 117.44 | 113.56 | 178.38 | 5 | 793 |
| 4 | dpp4_45_Evo_1 | 41.79 | 5.86 | 7.07 | 112.01 | 105.77 | 162.5 | 6 | 592 |
| 5 | dpp4_46_Evo_4 | 35.77 | 6.37 | 6.96 | 108.81 | 112.44 | 126.17 | 6 | 482 |
| 6 | dpp4_47_Evo_2 | 15.06 | 5.19 | 6.49 | 104.51 | 91.5 | 147.43 | 6 | 667 |
| 7 | dpp4_48_Evo_2 | 30.98 | 5.04 | 6.03 | 91.03 | 88.03 | 140.5 | 5 | 372 |
| 8 | dpp4_49_Evo_4 | 50.86 | 3.69 | 5.88 | 94.79 | 91.65 | 144.29 | 5 | 850 |
| 9 | dpp4_50_Evo_2 | 55.44 | 6.85 | 6.65 | 84.56 | 84.82 | 143.55 | 5 | 411 |
| 10 | dpp4_51_Evo_6 | 43.14 | 5.18 | 6.34 | 77.91 | 80.32 | 116.96 | 5 | 368 |
| 11 | Alogliptin_Evo_1 | 19.28 | 3.66 | 6.29 | 99.26 | 92.17 | 154.85 | 6 | 849 |
| Rank | Compounds from Databases | Fit Value | Predicted IC50 nM | Derivatives (de novo Evolution) | Fit Value | Predicted IC50 nM |
|---|---|---|---|---|---|---|
| 1 | dpp4_42 | 5.95 | 10.33 | dpp4_42_Evo_1 | 6.08 | 7.66 |
| 2 | dpp4_43 | 5.78 | 15.54 | dpp4_43_Evo_2 | 5.51 | 9.91 |
| 3 | dpp4_44 | 6.05 | 8.23 | dpp4_44_Evo_4 | 6.49 | 3.00 |
| 4 | dpp4_45 | 5.82 | 13.92 | dpp4_45_Evo_1 | 6.00 | 9.25 |
| 5 | dpp4_46 | 5.47 | 31.28 | dpp4_46_Evo_4 | 6.20 | 5.79 |
| 6 | dpp4_47 | 6.04 | 8.43 | dpp4_47_Evo_2 | 5.88 | 12.32 |
| 7 | dpp4_48 | 5.87 | 12.61 | dpp4_48_Evo_2 | 5.80 | 14.72 |
| 8 | dpp4_49 | 5.81 | 14.30 | dpp4_49_Evo_4 | 5.57 | 24.77 |
| 9 | dpp4_50 | 6.07 | 7.79 | dpp4_50_Evo_2 | 6.04 | 8.36 |
| 10 | dpp4_51 | 6.31 | 4.57 | dpp4_51_Evo_6 | 5.95 | 10.48 |
| 11 | Alogliptin | 5.62 | 22.18 | Alogliptin_Evo_1 | 5.56 | 25.26 |
| Ligand DPP-IV | 2i78_l | 3g0b_l | 5j3j_l | 5zid_l | 3vjk_ l | 5kby_l | 4n8d_l |
|---|---|---|---|---|---|---|---|
| 2i78 | 0.538 | 0.628 | 1.186 | 1.680 | 2.616 | 0.425 | 3.091 |
| 3g0b | 0.553 | 0.272 | 1.663 | 1.799 | 3.997 | 0.393 | 2.408 |
| 5j3j | 3.156 | 2.744 | 0.420 | 0.540 | 3.851 | 4.405 | 3.153 |
| 5zid | 2.928 | 2.192 | 0.465 | 1.596 | 2.336 | 2.438 | 2.303 |
| 3vjk | 0.601 | 2.229 | 0.801 | 0.836 | 0.934 | 4.272 | 2.256 |
| 5kby | 3.137 | 0.642 | 1.557 | 0.646 | 3.851 | 0.698 | 2.378 |
| 4n8d | 0.481 | 0.521 | 1.949 | 1.755 | 4.751 | 0.608 | 2.445 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musoev, A.; Numonov, S.; You, Z.; Gao, H. Discovery of Novel DPP-IV Inhibitors as Potential Candidates for the Treatment of Type 2 Diabetes Mellitus Predicted by 3D QSAR Pharmacophore Models, Molecular Docking and De Novo Evolution. Molecules 2019, 24, 2870. https://doi.org/10.3390/molecules24162870
Musoev A, Numonov S, You Z, Gao H. Discovery of Novel DPP-IV Inhibitors as Potential Candidates for the Treatment of Type 2 Diabetes Mellitus Predicted by 3D QSAR Pharmacophore Models, Molecular Docking and De Novo Evolution. Molecules. 2019; 24(16):2870. https://doi.org/10.3390/molecules24162870
Chicago/Turabian StyleMusoev, Azizullo, Sodik Numonov, Zhuhong You, and Hongwei Gao. 2019. "Discovery of Novel DPP-IV Inhibitors as Potential Candidates for the Treatment of Type 2 Diabetes Mellitus Predicted by 3D QSAR Pharmacophore Models, Molecular Docking and De Novo Evolution" Molecules 24, no. 16: 2870. https://doi.org/10.3390/molecules24162870
APA StyleMusoev, A., Numonov, S., You, Z., & Gao, H. (2019). Discovery of Novel DPP-IV Inhibitors as Potential Candidates for the Treatment of Type 2 Diabetes Mellitus Predicted by 3D QSAR Pharmacophore Models, Molecular Docking and De Novo Evolution. Molecules, 24(16), 2870. https://doi.org/10.3390/molecules24162870







