DMPK is a New Candidate Mediator of Tumor Suppressor p53-Dependent Cell Death
Abstract
:1. Introduction
2. Results
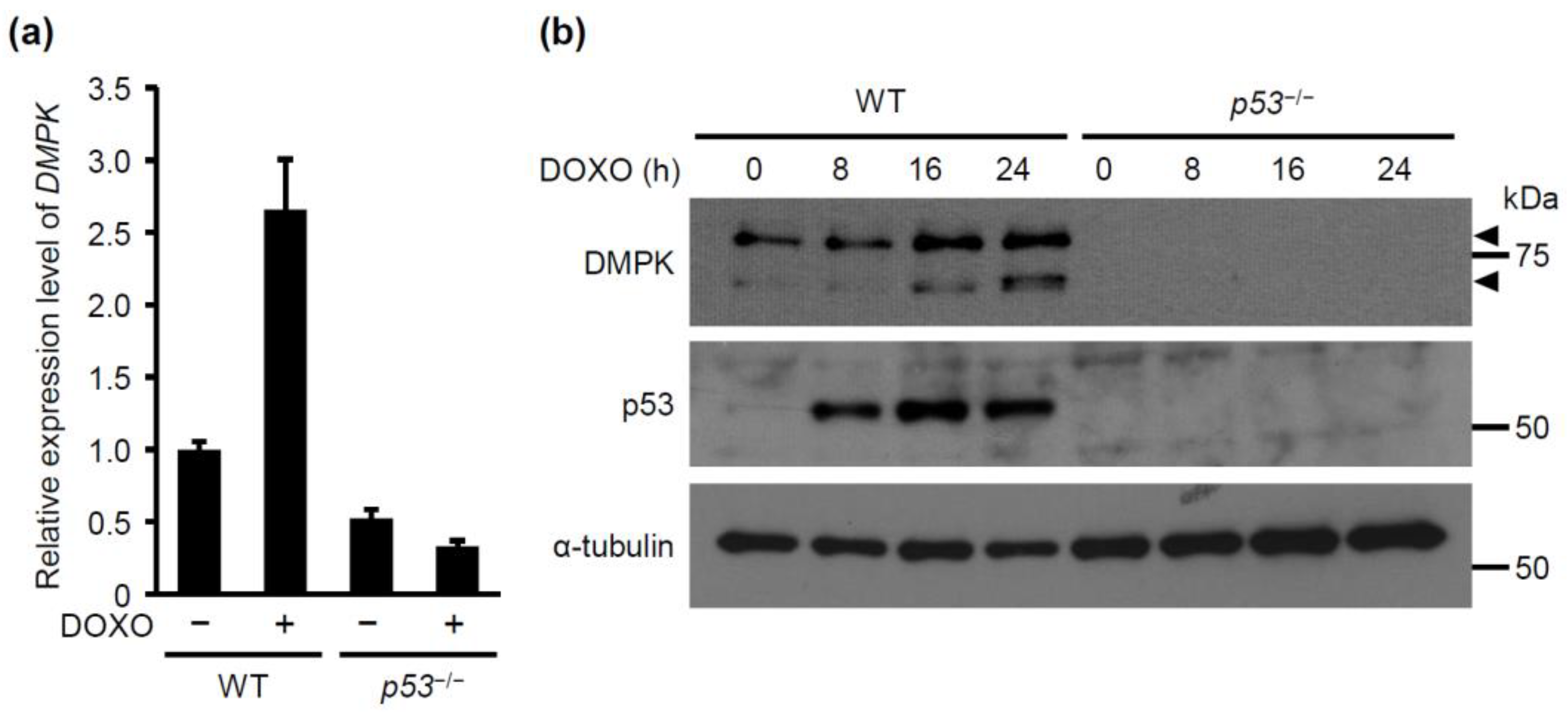
2.1. DMPK is Identified as a Novel p53 Downstream Target Gene Upon Doxorubicin Treatment
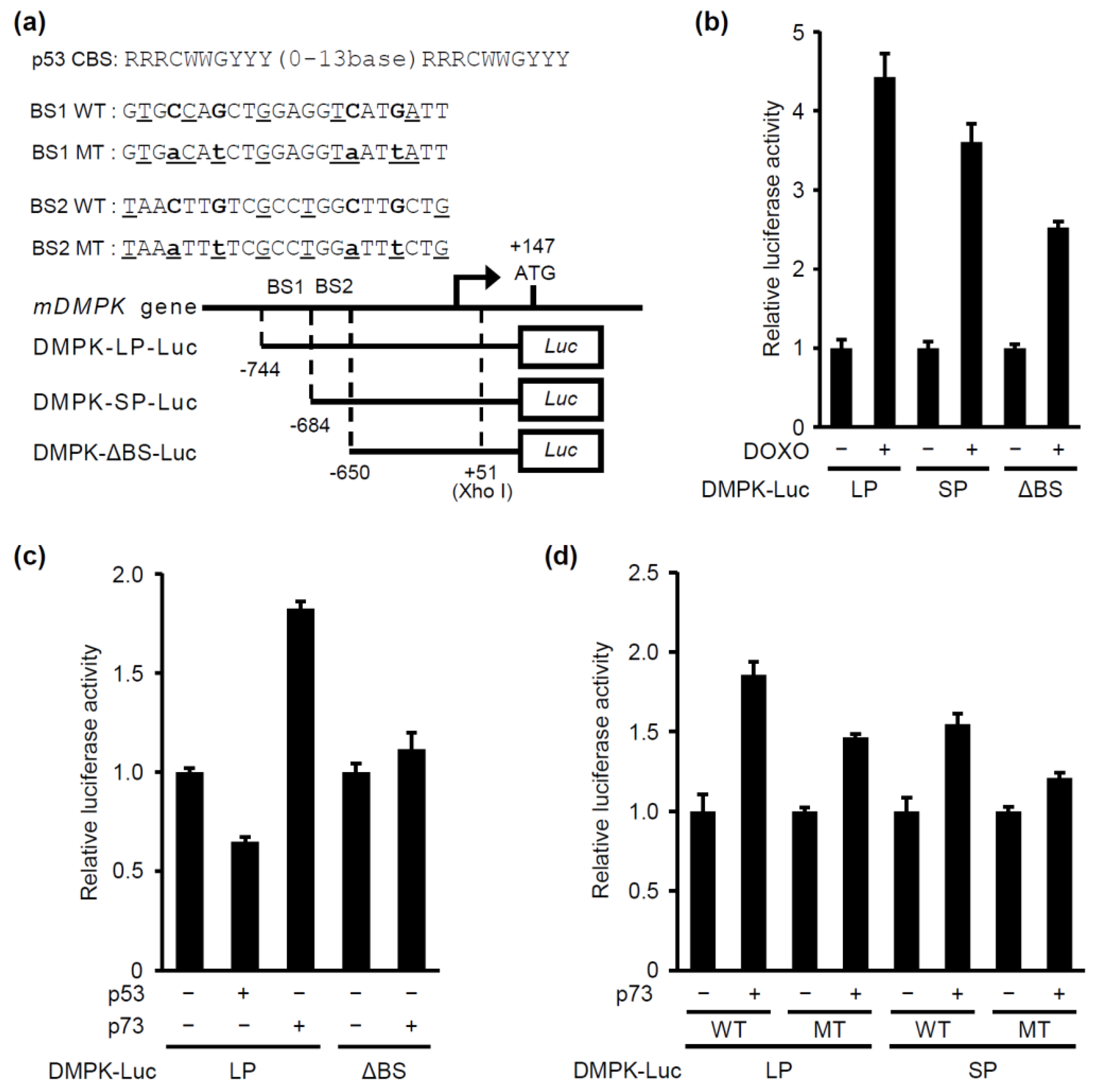
2.2. p73, but not p53, Induces DMPK Promoter Activation
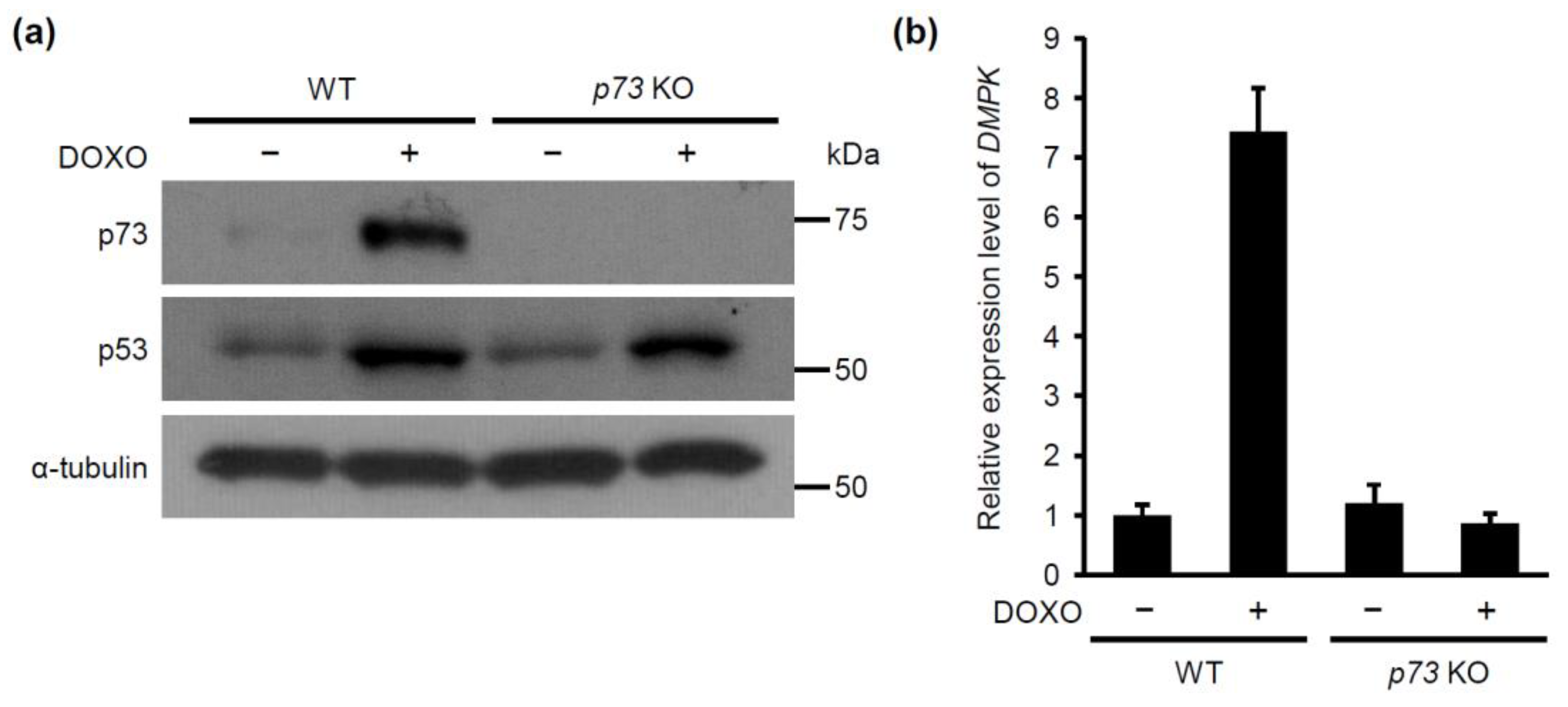
2.3. p53 is Required for Doxorubicin-Induced p73 Expression
2.4. p73 is Required for Doxorubicin-Induced DMPK Expression
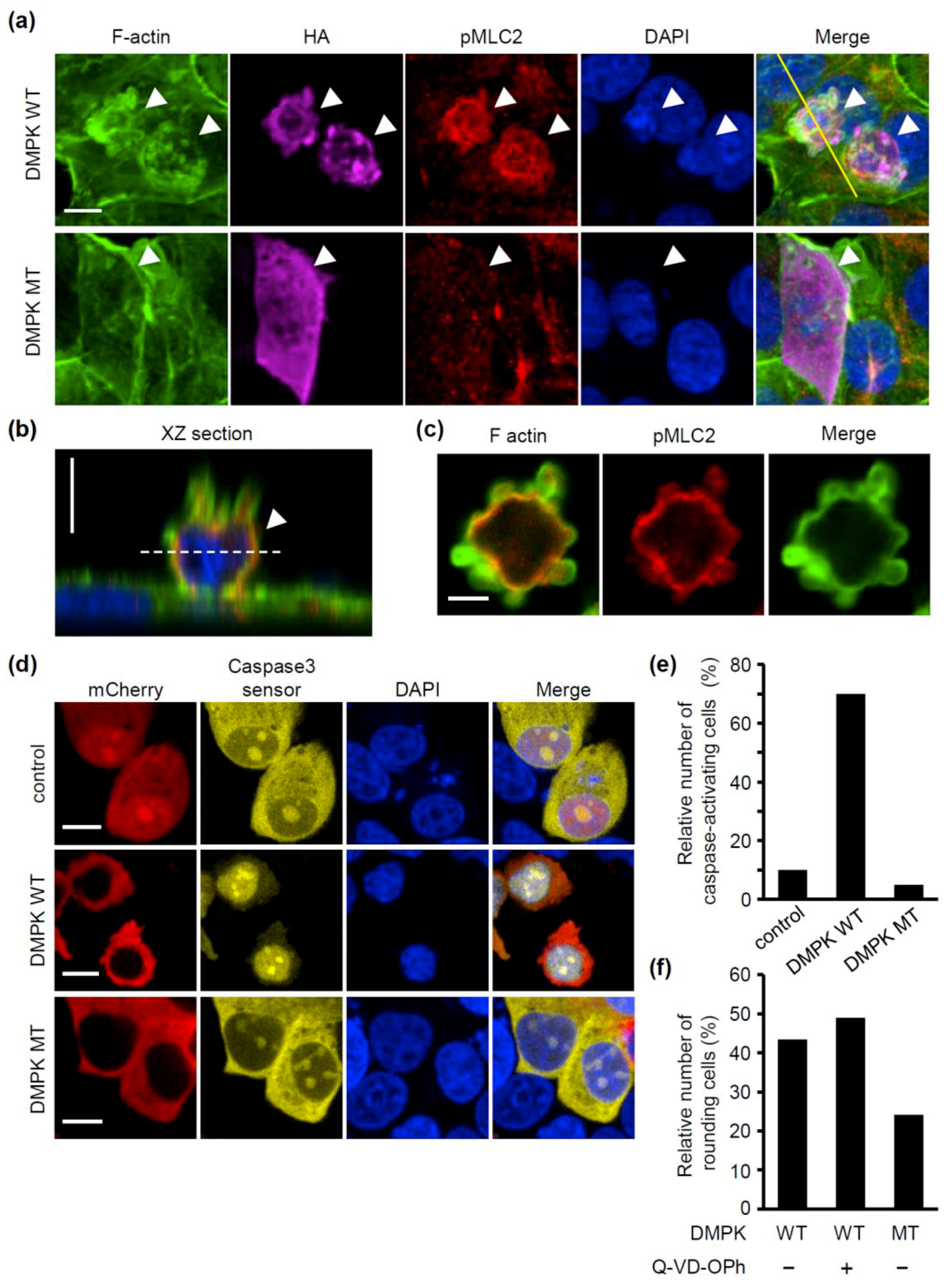
2.5. Overexpression of DMPK Induces Loss of Cell Adhesion and Caspase Activation
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Plasmidss
4.2. Antibodies and Materials
4.3. DNA Micro Array and Quantitative Real-Time PCR (qRT-PCR)
4.4. Immunoblot Analysis
4.5. Luciferase Assay
4.6. Fluorescence Microscopy
4.7. Evaluation of Caspase-Activation
4.8. Morphological Evaluation of Cells
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Desouza, M.; Gunning, P.W.; Stehn, J.R. The actin cytoskeleton as a sensor and mediator of apoptosis. Bioarchitecture 2012, 2, 75–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fackler, O.T.; Grosse, R. Cell motility through plasma membrane blebbing. J. Cell Biol. 2008, 181, 879–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vousden, K.H.; Lu, X. Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2002, 2, 594–604. [Google Scholar] [CrossRef]
- Kearns, S.; Lurz, R.; Orlova, E.V.; Okorokov, A.L. Two p53 tetramers bind one consensus DNA response element. Nucleic Acids Res. 2016, 44, 6185–6199. [Google Scholar] [CrossRef] [Green Version]
- Khoo, K.H.; Verma, C.S.; Lane, D.P. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014, 13, 217–236. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Vaseva, A.V.; Moll, U.M. The mitochondrial p53 pathway. Biochim. Biophys. Acta 2009, 1787, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Martinou, J.C.; Youle, R.J. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef]
- Melino, G.; De Laurenzi, V.; Vousden, K.H. p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer 2002, 2, 605–615. [Google Scholar] [CrossRef]
- Harms, K.; Nozell, S.; Chen, X. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 2004, 61, 822–842. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; De Laurenzi, V.; Munarriz, E.; Green, D.R.; Liu, Y.C.; Vousden, K.H.; Cesareni, G.; Melino, G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005, 24, 836–848. [Google Scholar] [CrossRef]
- Li, Y.; Prives, C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 2007, 26, 2220–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, K.; Fry, E.A. Alterations of p63 and p73 in human cancers. Subcell. Biochem. 2014, 85, 17–40. [Google Scholar] [PubMed]
- Vikhreva, P.; Melino, G.; Amelio, I. p73 Alternative Splicing: Exploring a Biological Role for the C-Terminal Isoforms. J. Mol. Biol. 2018, 430, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Pan, W.Y.; Chen, J. p53 and its isoforms in DNA double-stranded break repair. J. Zhejiang Univ. Sci. B 2019, 20, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Kaliman, P.; Llagostera, E. Myotonic dystrophy protein kinase (DMPK) and its role in the pathogenesis of myotonic dystrophy 1. Cell Signal. 2008, 20, 1935–1941. [Google Scholar] [CrossRef] [PubMed]
- Muranyi, A.; Zhang, R.; Liu, F.; Hirano, K.; Ito, M.; Epstein, H.F.; Hartshorne, D.J. Myotonic dystrophy protein kinase phosphorylates the myosin phosphatase targeting subunit and inhibits myosin phosphatase activity. FEBS Lett. 2001, 493, 80–84. [Google Scholar] [CrossRef]
- Wansink, D.G.; van Herpen, R.E.; Coerwinkel-Driessen, M.M.; Groenen, P.J.; Hemmings, B.A.; Wieringa, B. Alternative splicing controls myotonic dystrophy protein kinase structure, enzymatic activity, and subcellular localization. Mol. Cell. Biol. 2003, 23, 5489–5501. [Google Scholar] [CrossRef]
- Coleman, M.L.; Sahai, E.A.; Yeo, M.; Bosch, M.; Dewar, A.; Olson, M.F. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 2001, 3, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Sebbagh, M.; Renvoize, C.; Hamelin, J.; Riche, N.; Bertoglio, J.; Breard, J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 2001, 3, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Shimizu, M.; Balasubramanyam, A.; Epstein, H.F. Myotonic dystrophy protein kinase (DMPK) induces actin cytoskeletal reorganization and apoptotic-like blebbing in lens cells. Cell Motil. Cytoskelet. 2000, 45, 133–148. [Google Scholar] [CrossRef]
- Mulders, S.A.; van Horssen, R.; Gerrits, L.; Bennink, M.B.; Pluk, H.; de Boer-van Huizen, R.T.; Croes, H.J.; Wijers, M.; van de Loo, F.A.; Fransen, J.; et al. Abnormal actomyosin assembly in proliferating and differentiating myoblasts upon expression of a cytosolic DMPK isoform. Biochim. Biophys. Acta 2011, 1813, 867–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tophkhane, C.; Yang, S.H.; Jiang, Y.; Ma, Z.; Subramaniam, D.; Anant, S.; Yogosawa, S.; Sakai, T.; Liu, W.G.; Edgerton, S.; et al. p53 inactivation upregulates p73 expression through E2F-1 mediated transcription. PLoS ONE 2012, 7, e43564. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Washington, M.K.; Wei, J.; Chen, H.; Prassolov, V.S.; Zaika, A.I. Interactions of the p53 protein family in cellular stress response in gastrointestinal tumors. Mol. Cancer Ther. 2010, 9, 693–705. [Google Scholar] [CrossRef]
- Fridman, J.S.; Lowe, S.W. Control of apoptosis by p53. Oncogene 2003, 22, 9030–9040. [Google Scholar] [CrossRef] [Green Version]
- Freed-Pastor, W.A.; Prives, C. Mutant p53: One name, many proteins. Genes Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef]
- Hientz, K.; Mohr, A.; Bhakta-Guha, D.; Efferth, T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 2017, 8, 8921–8946. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef] [Green Version]
- Brook, J.D.; McCurrach, M.E.; Harley, H.G.; Buckler, A.J.; Church, D.; Aburatani, H.; Hunter, K.; Stanton, V.P.; Thirion, J.P.; Hudson, T.; et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell 1992, 68, 799–808. [Google Scholar] [CrossRef]
- Fu, Y.H.; Pizzuti, A.; Fenwick, R.G., Jr.; King, J.; Rajnarayan, S.; Dunne, P.W.; Dubel, J.; Nasser, G.A.; Ashizawa, T.; de Jong, P.; et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 1992, 255, 1256–1258. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.A. Molecular biology. Neutralizing toxic RNA. Science 2009, 325, 272–273. [Google Scholar] [PubMed]
- Savkur, R.S.; Philips, A.V.; Cooper, T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001, 29, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Guiraud-Dogan, C.; Huguet, A.; Gomes-Pereira, M.; Brisson, E.; Bassez, G.; Junien, C.; Gourdon, G. DM1 CTG expansions affect insulin receptor isoforms expression in various tissues of transgenic mice. Biochim. Biophys. Acta 2007, 1772, 1183–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, G.; Mahadevan, M.; Amemiya, C.; Wormskamp, N.; Segers, B.; Hendriks, W.; O’Hoy, K.; Baird, S.; Sabourin, L.; Lennon, G.; et al. Characterization of the myotonic dystrophy region predicts multiple protein isoform-encoding mRNAs. Nat. Genet. 1992, 1, 261–266. [Google Scholar] [CrossRef]
- Pham, Y.C.; Man, N.; Lam, L.T.; Morris, G.E. Localization of myotonic dystrophy protein kinase in human and rabbit tissues using a new panel of monoclonal antibodies. Hum. Mol. Genet. 1998, 7, 1957–1965. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, P.S.; Han, J.; Reddy, S. In situ hybridization analysis of Dmpk mRNA in adult mouse tissues. Neuromuscul. Disord. 2004, 14, 497–506. [Google Scholar] [CrossRef]
- Oude Ophuis, R.J.; Mulders, S.A.; van Herpen, R.E.; van de Vorstenbosch, R.; Wieringa, B.; Wansink, D.G. DMPK protein isoforms are differentially expressed in myogenic and neural cell lineages. Muscle Nerve 2009, 40, 545–555. [Google Scholar] [CrossRef]
- Kawauchi, K.; Araki, K.; Tobiume, K.; Tanaka, N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008, 10, 611–618. [Google Scholar] [CrossRef]
- Guo, A.K.; Hou, Y.Y.; Hirata, H.; Yamauchi, S.; Yip, A.K.; Chiam, K.H.; Tanaka, N.; Sawada, Y.; Kawauchi, K. Loss of p53 enhances NF-kappaB-dependent lamellipodia formation. J. Cell Physiol. 2014, 229, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, S.; Takeuchi, K.; Chihara, K.; Honjoh, C.; Kato, Y.; Yoshiki, H.; Hotta, H.; Sada, K. STAT1 is essential for the inhibition of hepatitis C virus replication by interferon-lambda but not by interferon-alpha. Sci. Rep. 2016, 6, 38336. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Data of the microarray is available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Itoh, K.; Ebata, T.; Hirata, H.; Torii, T.; Sugimoto, W.; Onodera, K.; Nakajima, W.; Uehara, I.; Okuzaki, D.; Yamauchi, S.; et al. DMPK is a New Candidate Mediator of Tumor Suppressor p53-Dependent Cell Death. Molecules 2019, 24, 3175. https://doi.org/10.3390/molecules24173175
Itoh K, Ebata T, Hirata H, Torii T, Sugimoto W, Onodera K, Nakajima W, Uehara I, Okuzaki D, Yamauchi S, et al. DMPK is a New Candidate Mediator of Tumor Suppressor p53-Dependent Cell Death. Molecules. 2019; 24(17):3175. https://doi.org/10.3390/molecules24173175
Chicago/Turabian StyleItoh, Katsuhiko, Takahiro Ebata, Hiroaki Hirata, Takeru Torii, Wataru Sugimoto, Keigo Onodera, Wataru Nakajima, Ikuno Uehara, Daisuke Okuzaki, Shota Yamauchi, and et al. 2019. "DMPK is a New Candidate Mediator of Tumor Suppressor p53-Dependent Cell Death" Molecules 24, no. 17: 3175. https://doi.org/10.3390/molecules24173175




