Drug Metabolite Cluster-Based Data-Mining Method for Comprehensive Metabolism Study of 5-hydroxy-6,7,3′,4′-tetramethoxyflavone in Rats
Abstract
:1. Introduction
2. Results
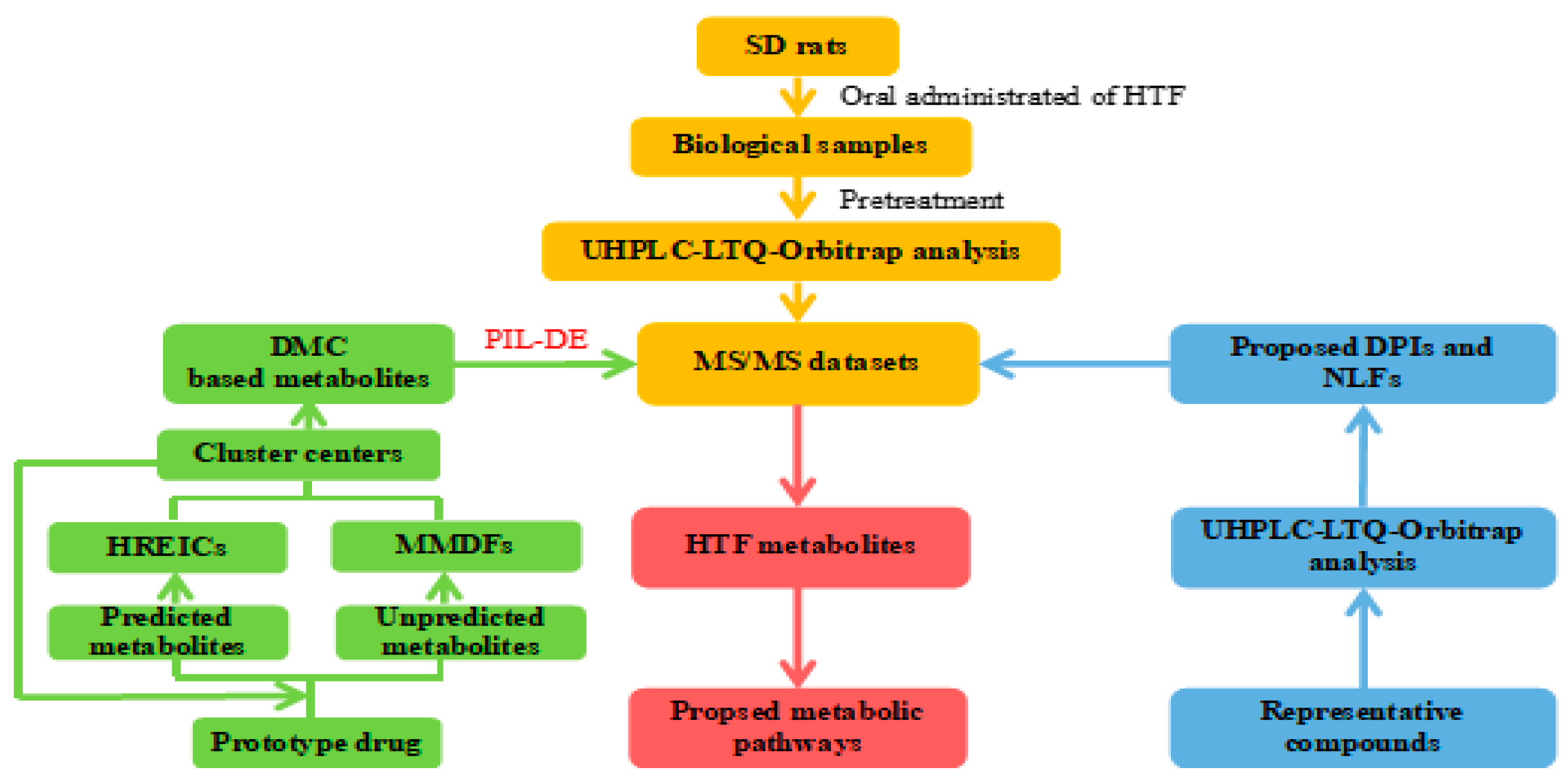
2.1. Establishment of HREIC and MMDF Based Data Processing Method
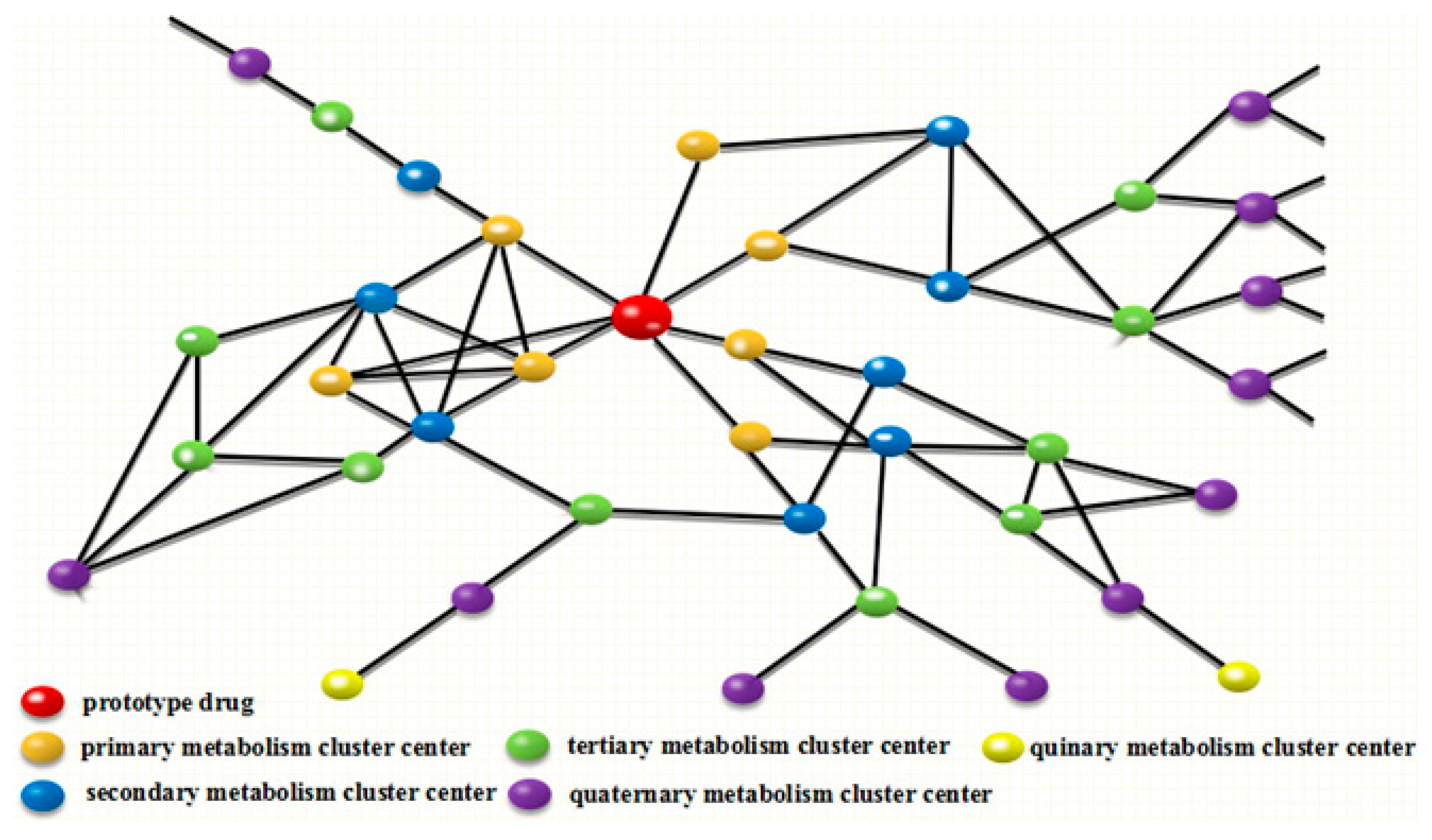
2.2. Establishment of DMC
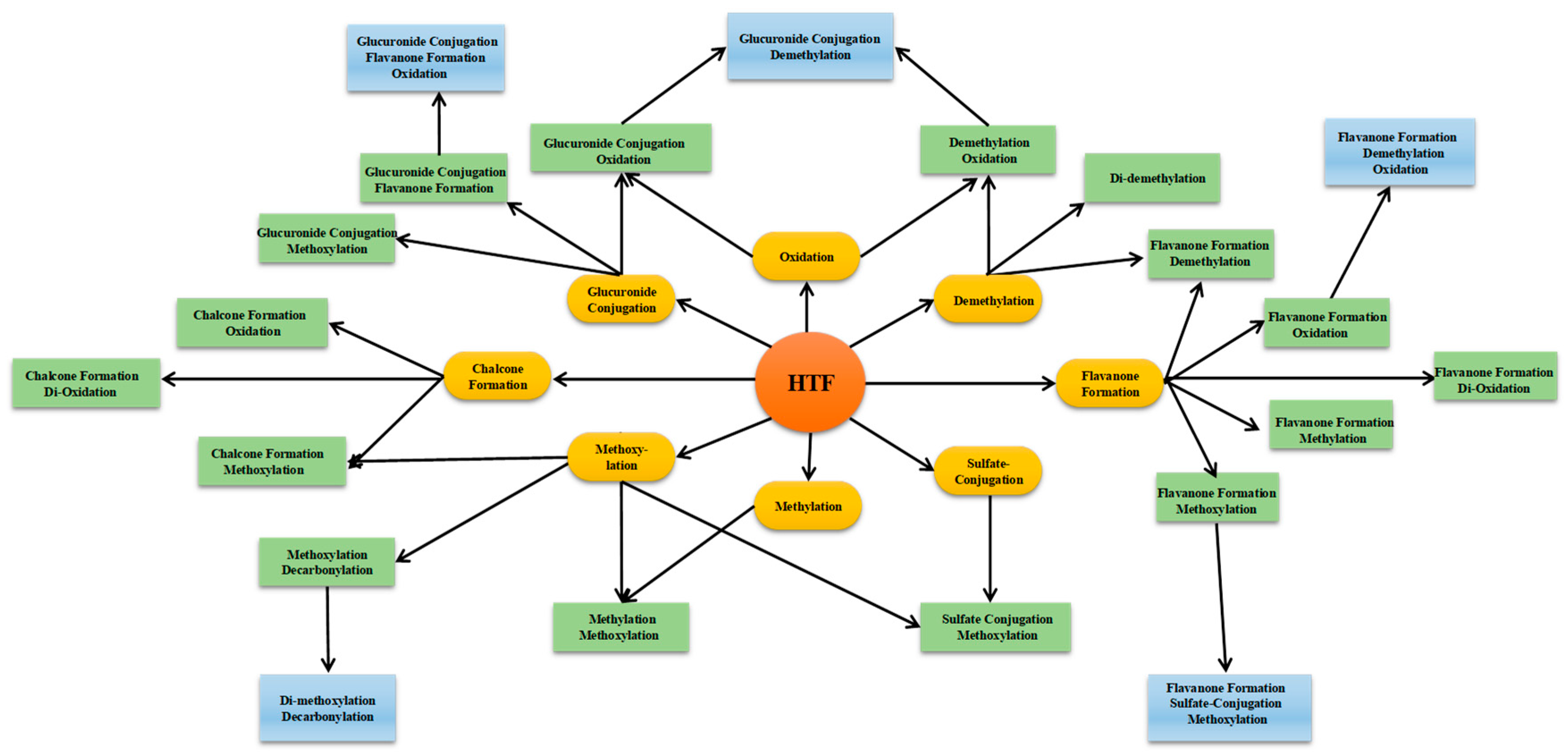
2.3. Combined Post-Acquisition Data Mining Methods
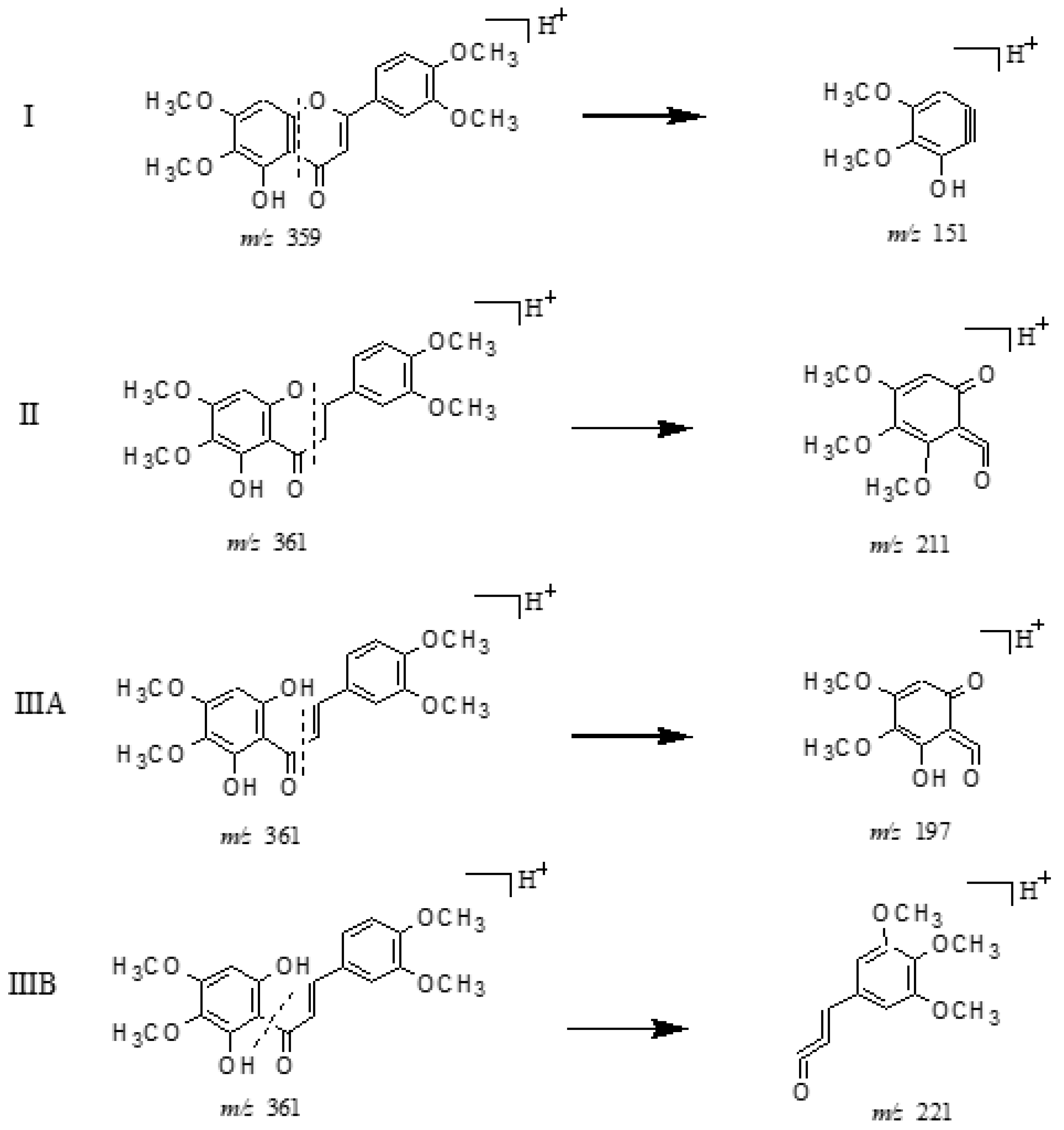
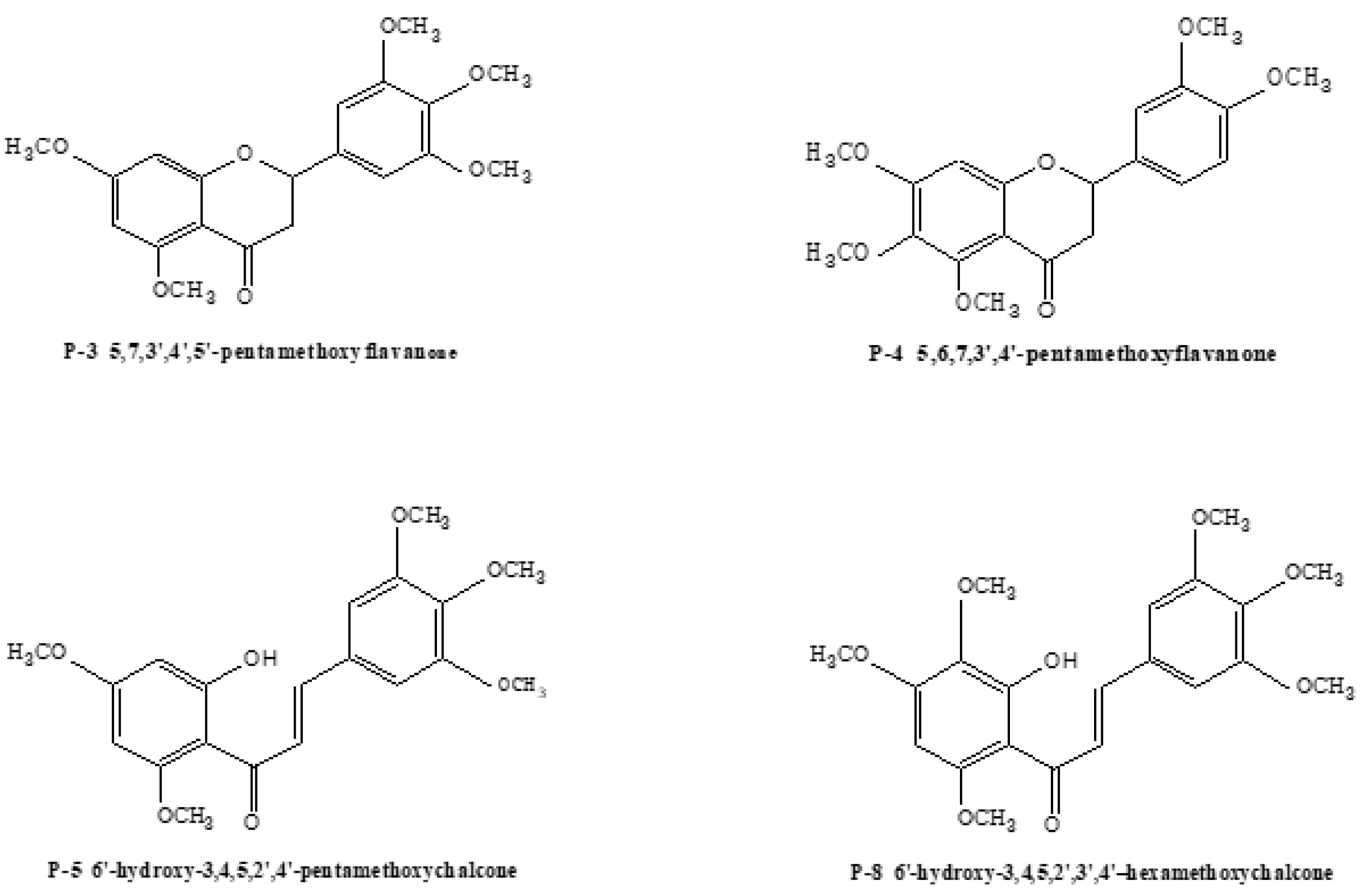
2.3.1. Structural Assignment of the Representative Polymethoxyflavones
2.3.2. Structural Assignment of the Representative Polymethoxyflavanones
2.3.3. Structural Assignment of the Representative Polymethoxychalcones
2.3.4. Structural Assignment of the Conjugate-Metabolites
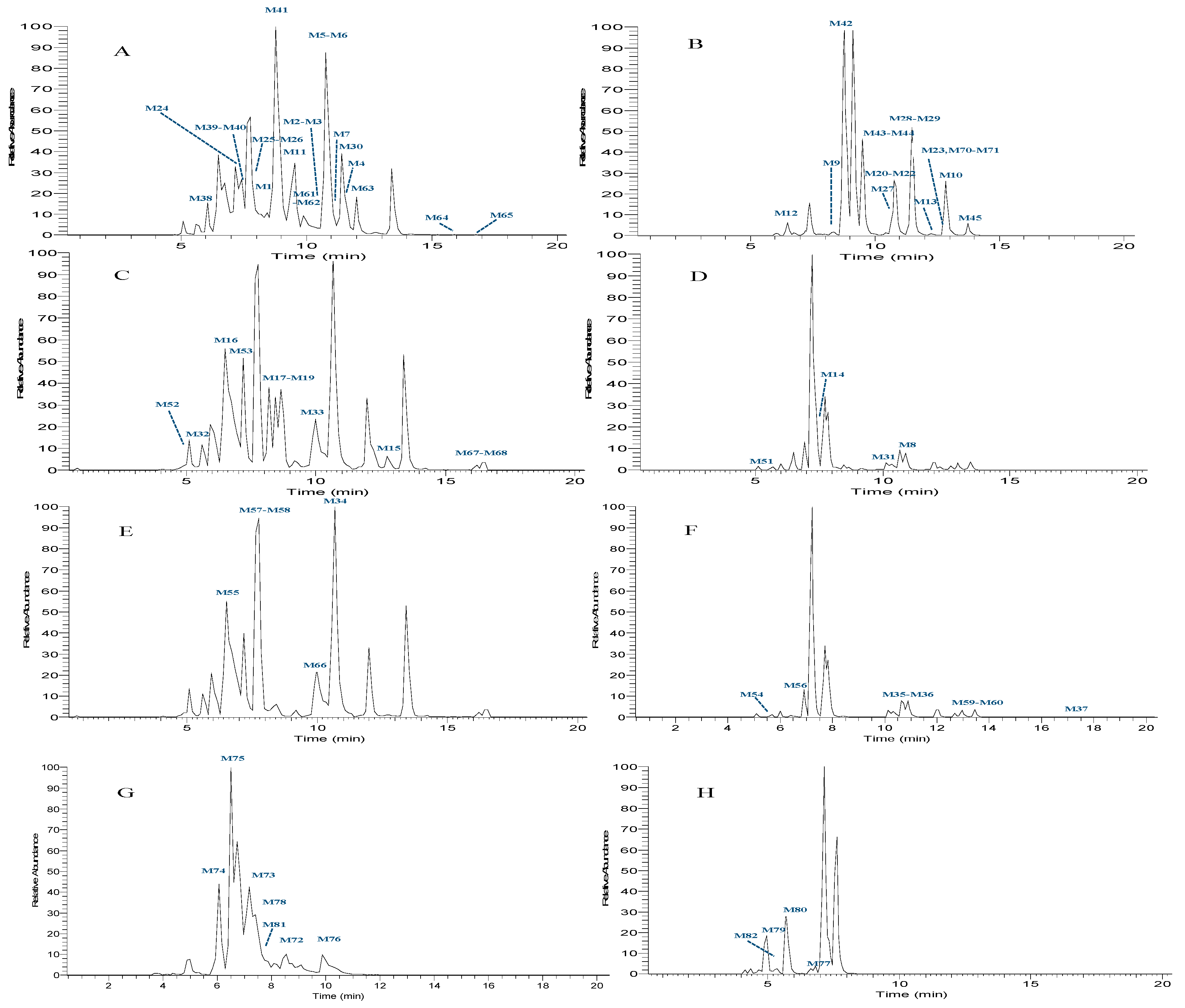
2.3.5. Summary of HTF Metablolites
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Animals and Drug Administration
4.3. Sample Collection and Preparation
4.4. Instrument and Conditions
4.5. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Trifonova, O.; Knight, R.A.; Lisitsa, A.; Melino, G.; Antonov, A.V.J.D.D.T. Exploration of individuality in drug metabolism by high-throughput metabolomics: The fast line for personalized medicine. Drug Discov. Today 2016, 21, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cai, W.; Cai, Q.; Che, Y.; Zhao, B.; Zhang, J. Comprehensive characterization of the in vitro and in vivo metabolites of geniposide in rats using ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer. Xenobiotica 2015, 46, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, S.; Li, L.P.; Jiang, H. Metabolism of flavonoids in human: A comprehensive review. Curr. Drug Metab. 2014, 15, 1. [Google Scholar] [CrossRef]
- Sandlers, Y.J.T.R. The future perspective: Metabolomics in laboratory medicine for inborn errors of metabolism. Transl. Res. 2017, 189, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Xu, B.; Wang, Y.; Shi, X.; Qiao, Y.; Zhang, J. Metabolomics data fusion between near infrared spectroscopy and high-resolution mass spectrometry: A synergetic approach to boost performance or induce confusion. Talanta 2018, 189, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Dictionary of Natural Products. Available online: http://dnp.chemnetbase.com (accessed on 10 August 2009).
- Miller, M.J.; Kennedy, A.D.; Eckhart, A.D.; Burrage, L.C.; Wulff, J.E.; Miller, L.A.D.; Milburn, M.V.; Ryals, J.A.; Beaudet, A.L.; Sun, Q.J.J.; et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. Inherit. Metab. Dis. 2015, 38, 1029–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Donti, T.R.; Cappuccio, G.; Hubert, L.; Neira, J.; Atwal, P.S.; Miller, M.J.; Cardon, A.L.; Sutton, V.R.; Porter, B.E.; Baumer, F.M. Diagnosis of adenylosuccinate lyase deficiency by metabolomic profiling in plasma reveals a phenotypic spectrum. Mol. Genet. Metab. Rep. 2016, 8, 61–66. [Google Scholar] [CrossRef]
- Coene, K.L.M.; Kluijtmans, L.A.J.; Heeft, E.V.D.; Engelke, U.F.H.; Boer, S.D.; Hoegen, B.; Kwast, H.J.T.; Vorst, M.V.D.; Huigen, M.C.D.G.; Keularts, I.M.L.W. Next-generation metabolic screening: Targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J. Inherit. Metab. Dis. 2018, 41, 337–353. [Google Scholar] [CrossRef]
- Farid, N.A.; Kurihara, A.; Wrighton, S.A. Metabolism and Disposition of the Thienopyridine Antiplatelet Drugs Ticlopidine, Clopidogrel, and Prasugrel in Humans. Clin. Pharmacol. 2013, 50, 126–142. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006, 8, E101–E111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.Y.; Yang, L.; Wei, J.; Huang, M.; Jiang, J.G. Bioactivity evaluations of ingredients extracted from the flowers of Citrus aurantium L. var. amara Engl. Food Chem. 2012, 135, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Nakata, A.; Koike, Y.; Matsui, H.; Shimadad, T.; Aburada, M.; Yang, J. Potent SIRT1 enzyme-stimulating and anti-glycation activities of polymethoxyflavonoids from Kaempferia parviflora. Nat. Prod. Commun. 2014, 9, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, I.N.; Ho, C.T.; Li, S.; Colby, J.; Dushenkov, S. Apoptosis-inducing activity of hydroxylated polymethoxyflavones and polymethoxyflavones from orange peel in human breast cancer cells. Mol. Nutr. Food Res. 2010, 51, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.S.; Li, W.; Li, Y.; Wang, Q. Synthesis of Citrus polymethoxyflavonoids and their antiproliferative activities on Hela cells. Med. Chem. Res. 2017, 26, 1–8. [Google Scholar] [CrossRef]
- Tominari, T.; Hirata, M.; Matsumoto, C.; Inada, M.; Miyaura, C. Polymethoxy Flavonoids, Nobiletin and Tangeretin, Prevent Lipopolysaccharide-Induced Inflammatory Bone Loss in an Experimental Model for Periodontitis. Pharmacol. Sci. 2012, 119, 390–394. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.M.; Tait, A.R.; Kitts, D.D. Flavonoid composition of orange peel and its association with antioxidant and anti-inflammatory activities. Food Chem. 2017, 218, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A.; Nakamura, Y.; Ohto, Y.; Yano, M.; Koshiba, T.; Koshimizu, K.; Tokuda, H.; Nishino, H.; Ohigashi, H. Suppressive effects of citrus fruits on free radical generation and nobiletin, an anti-inflammatory polymethoxyflavonoid. Biofactors 2010, 12, 187–192. [Google Scholar] [CrossRef]
- Li, S.; Pan, M.H.; Lo, C.Y.; Tan, D.; Wang, Y.; Shahidi, F.; Ho, C.T. Chemistry and health effects of polymethoxyflavones and hydroxylated polymethoxyflavones. Funct. Foods 2009, 1, 2–12. [Google Scholar] [CrossRef]
- Yoshizaki, N.; Fujii, T.; Masaki, H.; Okubo, T.; Shimada, K.; Hashizume, R. Orange peel extract, containing high levels of polymethoxyflavonoid, suppressed UVB-induced COX-2 expression and PGE2 production in HaCaT cells through PPAR-γ activation. Exp. Dermatol. 2015, 23, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Cao, A.; Shi, J.; Yin, P.; Wang, L.; Ji, G.; Xie, J.; Wu, D. Tangeretin, a citrus polymethoxyflavonoid, induces apoptosis of human gastric cancer AGS cells through extrinsic and intrinsic signaling pathways. Oncol. Rep. 2014, 31, 1788–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bingol, K.; Bruschweiler-Li, L.; Li, D.; Zhang, B.; Xie, M.; Brüschweiler, R. Emerging new strategies for successful metabolite identification in metabolomics. Bioanalysis 2016, 8, 557–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Zhong, D.; Yu, K.; Chen, X. Recent advances in metabolite identification and quantitative bioanalysis by LC–Q-TOF MS. Bioanalysis 2012, 4, 937–959. [Google Scholar] [CrossRef] [PubMed]
- Bouslimani, A.; Sanchez, L.M.; Garg, N.; Dorrestein, P.C. Mass spectrometry of natural products: Current, emerging and future technologies. Nat. Prod. Rep. 2014, 31, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Cai, W.; Cao, Y.; Wang, F.; Wang, Z.; Lu, J.; Zhang, J. An integrated strategy for rapid discovery and identification of the sequential piperine metabolites in rats using ultra high-performance liquid chromatography/high resolution mass spectrometery. J. Pharm. Biomed. Anal. 2017, 146, 387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, D.; Ray, K.; Zhu, M. Mass defect filter technique and its applications to drug metabolite identification by high-resolution mass spectrometry. J. Mass Spectrom. 2010, 44, 999–1016. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, L.; Leng, J.; Chen, P.; Fan, X.; Cheng, Y. Fragment ion diagnostic strategies for the comprehensive identification of chemical profile of Gui-Zhi-Tang by integrating high-resolution MS, multiple-stage MS and UV information. Pharm. Biomed. Anal. 2014, 98, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Wang, Z.J.; Zhang, Q.; Wang, F.; Ma, Q.; Lin, Z.Z.; Lu, J.Q.; Qiao, Y.J. Rapid screening and identification of target constituents using full scan-parent ions list-dynamic exclusion acquisition coupled to diagnostic product ions analysis on a hybrid LTQ-Orbitrap mass spectrometer. Talanta 2014, 124, 111–122. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Wang, Z.J.; Li, Y.; Liu, Y.; Cai, W.; Li, C.; Lu, J.Q.; Qiao, Y.J. A strategy for comprehensive identification of sequential constituents using ultra–High–performance liquid chromatography coupled with linear ion trap–Orbitrap mass spectrometer, application study on chlorogenic acids in Flos Lonicerae Japonicae. Talanta 2016, 147, 16–27. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| No. | Formula [M + H]+ | Theoretical Mass m/z | Experimental Mass m/z | Error (ppm) | MS/MS Fragment Ions |
|---|---|---|---|---|---|
| P-1 | C19H19O8 | 359.11256 | 359.11290 | 1.0 | MS2[359]:326(100),344(71) |
| MS3[326]:298(100),278(0.6) | |||||
| P-2 | C18H17O6 | 329.10196 | 329.10123 | −2.2 | MS2[329]:314(100),313(66),285(21),268(4),192(0.5) |
| MS3[314]:283(100),298(37),167(24),270(24),173(2) | |||||
| P-3 | C20 H19O7 | 375.12817 | 375.12839 | 0.5 | MS2[375]:211(100),191(18),357(13) |
| MS3[211]:196(100),183(29),178(29),150(22) | |||||
| P-4 | C20H23O7 | 375.14382 | 375.14420 | 1.0 | MS2[375]:211(100),191(43),357(20) |
| MS3[211]:196(100),178(24),183(9) | |||||
| P-5 | C20H23O7 | 375.14382 | 375.14420 | 1.0 | MS2[375]:221(100),181(9) |
| MS3[221]:193(100),190(51),191(37),206(26) | |||||
| P-6 | C20 H21O8 | 389.12309 | 389.12289 | −0.3 | MS2[389]:359(100),328(70),374(66),356(42),328(15) |
| MS3[359]:344(100),341(72),343(66),191(39) | |||||
| P-7 | C20 H21O8 | 389.12309 | 389.12271 | −0.9 | MS2[389]:374(100),328(95),359(43),356(14) |
| MS3[374]:345(100),312(81),358(29),341(26) | |||||
| P-8 | C21H25O8 | 405.15439 | 405.15430 | −0.2 | MS2[405]:221(100),387(26),211(17) |
| MS3[221]:193(100),190(63),191(39),206(15) | |||||
| P-9 | C20 H21O8 | 419.13365 | 419.13360 | −0.1 | MS2[419]:389(100),404(66) |
| MS3[389]:356(100),371(90),361(85),359(79),374(79) |
| Peak | Ion Mode | tR /min | Formula | Theoretical Mass m/z | Experimental Mass m/z | Error (ppm) | MS/MS Fragment Ions | Identification/Reactions |
|---|---|---|---|---|---|---|---|---|
| M0 | N | 8.86 | C19H17O7 | 357.09692 | 357.09698 | 0.2 | MS2[357]:342(100),327(7),314(2),283(2),284(1),282(0.6),324(0.4) | HTF |
| MS3[342]:327(100),312(4),299(1),324(1),314(0.6),296(0.5) | ||||||||
| P | 8.86 | C19H19O7 | 359.11256 | 359.11218 | −0.9 | MS2[359]:298(100),344(71),316(28),329(24),326(22),343(14) | ||
| MS3[298]:283(100),255(18),297(17),270(16),282(14),151(2) | ||||||||
| M1 | N | 7.94 | C17H13O7 | 329.06552 | 329.06497 | −1.8 | MS2[329]:314(100),299(6),285(1),315(0.6) | Loss of 2CH2 |
| MS3[314]:299(100),313(49),285(47),241(4),286(3),296(3) | ||||||||
| M2 | N | 10.01 | C17H13O7 | 329.06552 | 329.06628 | 2.1 | MS2[329]:229(100),211(76),311(29),293(24),171(21) | Loss of 2CH2 |
| MS3[229]:211(100),209(54),125(21),155(18),167(15),127(12) | ||||||||
| M3 | N | 10.24 | C17H13O7 | 329.06552 | 329.06714 | 4.7 | MS2[329]:171(100),229(56),293(54),311(49),211(40), | Loss of 2CH2 |
| MS3[171]:127(100),153(74),125(37) | ||||||||
| M4 | N | 11.65 | C17H13O7 | 329.06552 | 329.06677 | 3.6 | MS2[329]:171(100),201(77),311(56),275(40),293(37),213(7),185(4) | Loss of 2CH2 |
| MS3[171]:127(100),153(71),125(39) | ||||||||
| M5 | N | 10.77 | C18H15O7 | 343.08117 | 343.08008 | −3.3 | MS2[343]:328(100),313(3) | Loss of CH2 |
| MS3[328]:313(100),312(25),299(4),300(2),151(1) | ||||||||
| P | 10.77 | C18H17O7 | 345.09687 | 345.09579 | −3.1 | MS2[345]:330(100),312(96),284(5),327(1) | ||
| M6 | N | 10.97 | C18H15O7 | 343.08117 | 343.08066 | −3.3 | MS2[343]:328(100),329(6),313(5),315(1) | Loss of CH2 |
| MS3[328]:313(100),312(10),299(3),300(2),285(1),298(1) | ||||||||
| P | 10.97 | C18H17O7 | 345.09687 | 345.09558 | −3.7 | MS2[345]:312(100),330(93),284(5),327(1) | ||
| M7 | N | 11.26 | C18H15O7 | 343.08117 | 343.08118 | −0.1 | MS2[343]:328(100),313(5),315(1) | Loss of CH2 |
| MS3[328]:313(100),312(15),282(14),309(10),299(7),300(2),151(1) | ||||||||
| P | 11.26 | C18H17O7 | 345.09687 | 345.09546 | −4.1 | MS2[345]:330(100),312(94),284(18),315(3),329(3),327(1) | ||
| M8 | P | 10.65 | C18H19O7 | 347.11247 | 347.11169 | −2.4 | MS2[347]:197(100),177(16),145(4),305(3),182(3),348(2),223(2) | Loss of CH2 + Flavanone Formation |
| MS3[197]:182(100),164(19),165(5),136(3),137(2) | ||||||||
| M9 | P | 8.37 | C19H19O7 | 359.11256 | 359.11200 | −1.4 | MS2[359]:344(100),326(67),343(2),298(2),327(1) | HTF isomer |
| MS3[344]:326(100),329(2),298(2),315(1) | ||||||||
| M10 | P | 12.83 | C19H19O7 | 359.11256 | 359.11142 | −3.0 | MS2[359]:326(100),344(73),343(8),298(5),327(2) | HTF isomer |
| MS3[326]:298(100),299(1) | ||||||||
| M11 | N | 8.97 | C18H15O8 | 359.07721 | 359.0755 | −1.7 | MS2[359]:344(100),343(9),329(1) | Loss of CH2 + Oxidation |
| MS3[344]:329(100),328(21),315(4),316(2),314(2) | ||||||||
| M12 | P | 6.40 | C19H21O7 | 361.12817 | 361.12750 | −1.8 | MS2[361]:197(100),191(40),343(25),329(9),301(6),182(3) | Loss of Carbonyl + Methoxylation |
| MS3[197]:182(100),165(18),123(5),169(5),137(4),151(4) | ||||||||
| M13 | P | 12.57 | C19H21O7 | 361.12817 | 361.12738 | −2.2 | MS2[361]:197(100),191(39),211(38),343(9),223(7),177(6),329(4) | Loss of Carbonyl + Methoxylation |
| MS3[197]:182(100),165(17),164(9),169(8),151(2) | ||||||||
| M14 | P | 7.33 | C19H21O7 | 361.12817 | 361.12769 | −1.3 | MS2[361]:211(100),177(16),329(6),343(6),145(4),196(4) | Flavanone Formation |
| MS3[211]:196(100),178(22),150(5),151(3),183(2),179(1) | ||||||||
| M15 | N | 12.23 | C19H19O7 | 359.11362 | 359.11261 | 0.2 | MS2[359]:149(100),134(35),344(20),343(2),329(2),175(2),179(1) | Flavanone Formation |
| MS3[149]:134(100) | ||||||||
| P | 12.25 | C19H21O7 | 361.12817 | 361.12769 | −1.9 | MS2[361]:211(100),177(16),343(7),329(6),145(4),196(4) | ||
| MS3[211]:196(100),178(25),150(5),183(4),151(3) | ||||||||
| M16 | N | 6.40 | C18H17O8 | 361.09182 | 361.09155 | −0.6 | MS2[361]:346(100),343(9),181(6),166(4),207(2) | Loss of CH2 + Oxidation and Flavanone Formation |
| MS3[346]:180(100),166(77),290(54),328(50),331(43),152(26) | ||||||||
| M17 | N | 8.20 | C18H17O8 | 361.09182 | 361.09155 | −0.6 | MS2[361]:346(100),343(9),181(6),166(4),207(2),328(1) | Loss of CH2 + Oxidation and Flavanone Formation |
| MS3[346]:180(100),166(77),290(54),328(50),331(43),318(38) | ||||||||
| M18 | N | 8.42 | C18H17O8 | 361.09182 | 361.09152 | −0.7 | MS2[361]:195(100),346(43),180(41),165(37),150(13),343(12),191(5) | Loss of CH2 + Oxidation and Flavanone Formation |
| MS3[195]:180(100),127(1) | ||||||||
| P | 8.46 | C18H19O8 | 363.10744 | 363.1066 | −2.3 | MS2[363]:193(100),197(94),345(51),161(23),133(15),364(7),182(2) | ||
| MS3[193]:161(100),133(14),178(2),165(1) | ||||||||
| M19 | N | 8.65 | C18H17O8 | 361.09182 | 361.09137 | −1.1 | MS2[361]:346(100),195(48),180(19),165(17),362(11),150(6) | Loss of CH2 + Oxidation and Flavanone Formation |
| MS3[346]:331(100),166(14),328(12),313(4),297(4),303(3) | ||||||||
| M20 | P | 10.64 | C20H21O7 | 373.12822 | 373.12738 | −2.1 | MS2[373]:312(100),358(62),329(30),343(27),340(22),357(18),339(3) | Methylation |
| MS3[312]:297(100),296(79),151(42),269(36),268(35),281(17),311(15) | ||||||||
| M21 | P | 10.76 | C20H21O7 | 373.12822 | 373.12726 | −2.4 | MS2[373]:312(100),358(69),329(28),343(27),340(22),357(18),339(3) | Methylation |
| MS3[312]:297(100),268(53),296(46),269(45),151(40),284(17),311(15) | ||||||||
| M22 | P | 10.92 | C20H21O7 | 373.12822 | 373.12701 | −3.1 | MS2[373]:312(100),358(73),329(28),343(27),340(20),357(15),313(12) | Methylation |
| M23 | P | 12.71 | C20H21O7 | 373.12822 | 373.12619 | −4.3 | MS2[373]:355(100),337(8),319(4) | Methylation |
| MS3[355]:337(100),319(60),227(50),213(41),241(32),145(31),173(20) | ||||||||
| M24 | N | 7.42 | C19H17O8 | 373.09172 | 373.09167 | −0.3 | MS2[373]:358(100),357(7),343(6) | Oxidation |
| MS3[358]:343(100),328(14),330(5),340(1),315(1),329(1) | ||||||||
| M25 | N | 8.02 | C19H17O8 | 373.09172 | 373.09094 | −2.2 | MS2[373]:358(100),343(6),357(2),313(1),269(1),295(1) | Oxidation |
| MS3[358]:343(100),330(2),327(1) | ||||||||
| M26 | N | 8.61 | C19H17O8 | 373.09172 | 373.08969 | −4.6 | MS2[373]:358(100),313(1),357(1) | Oxidation |
| MS3[358]:343(100),330(8),329(1) | ||||||||
| M27 | P | 10.44 | C19H19O8 | 375.10737 | 375.1066 | −2.2 | MS2[375]:314(100),345(24),360(13),342(12) | Oxidation |
| MS3[314]:286(100),285(42),299(5),287(4) | ||||||||
| M28 | N | 10.71 | C19H17O8 | 373.09172 | 373.09152 | −0.7 | MS2[373]:358(100),98(4),175(4),190(4),124(4) | Oxidation |
| P | 10.71 | C19H19O8 | 375.10737 | 375.10645 | −2.6 | MS2[375]:342(100),360(74),375(12),314(10) | ||
| MS3[342]:314(100),315(0.6) | ||||||||
| M29 | N | 11.38 | C19H17O8 | 373.09172 | 373.09094 | −2.2 | MS2[373]:358(100),374(26),373(6),359(5),343(3) | Oxidation |
| MS3[358]:343(100),329(1),341(0.5) | ||||||||
| P | 11.37 | C19H19O8 | 375.10737 | 375.10641 | −2.7 | MS2[375]:360(100),376(38),359(38),342(36),375(29),314(11) | ||
| MS3[360]:342(100),326(64),344(48),359(46),314(29),327(15),331(10),345(10) | ||||||||
| M30 | N | 11.83 | C19H17O8 | 373.09172 | 373.09195 | 0.4 | MS2[373]:358(100),343(3),357(1),359(1) | Oxidation |
| MS3[358]:343(100),342(7),328(2),299(1),284(1) | ||||||||
| M31 | P | 9.19 | C20H23O7 | 375.14327 | 375.14301 | −2.1 | MS2[375]:211(100),191(43),357(23),376(22),343(7) | Methylation + Flavanone Formation |
| MS3[211]:196(100),178(24),183(19),151(4),179(2) | ||||||||
| M32 | N | 5.94 | C19H19O8 | 375.10747 | 375.10773 | 0.7 | MS2[375]:165(100),150(39),360(10),191(3),325(1) | Oxidation + Flavanone Formation |
| MS3[165]:150(100),123(0.2),121(0.1) | ||||||||
| P | 6.00 | C19H21O8 | 377.12312 | 377.12222 | −2.3 | MS2[377]:359(100),193(73),211(67),161(19),133(14) | ||
| MS3[359]:300(100),299(95),327(67),328(52),344(36),331(35),313(29) | ||||||||
| M33 | P | 10.15 | C19H21O8 | 377.12312 | 377.12198 | −2.9 | MS2[377]:207(100),197(75),359(29),175(23),159(17),182(2) | Oxidation + Flavanone Formation |
| MS3[207]:175(100),145(7),192(2),119(2) | ||||||||
| M34 | N | 10.67 | C19H19O8 | 375.10747 | 375.10703 | −1.1 | MS2[375]:360(100),345(4),361(4),166(2) | Oxidation + Chalcone Formation |
| MS3[360]:345(100),166(46),332(14),179(11),304(11),317(10) | ||||||||
| P | 10.67 | C19H21O8 | 377.12312 | 377.12186 | −3.2 | MS2[377]:221(100),209(38),183(28),343(19),361(17),359(11) | ||
| MS3[221]:193(100),150(58),206(45),191(42),189(19),178(5) | ||||||||
| M35 | P | 10.98 | C19H21O8 | 377.12312 | 377.12225 | −2.2 | MS2[377]:197(100),207(82),359(33),175(21),221(14) | Oxidation + Chalcone Formation |
| MS3[197]:182(100),164(21),165(5),137(3) | ||||||||
| M36 | P | 11.14 | C19H21O8 | 377.12312 | 377.12173 | −3.6 | MS2[377]:197(100),207(84),221(54),359(34),209(19),175(14),183(14) | Oxidation + Chalcone Formation |
| MS3[197]:182(100),164(21),165(9),137(3),153(2) | ||||||||
| M37 | P | 17.02 | C19H21O8 | 377.12312 | 377.12207 | −2.7 | MS2[377]:359(100),291(99),289(20),221(19),207(17),197(17),175(3) | Oxidation + Chalcone Formation |
| MS3[359]:327(100),267(63),341(51),331(45),278(30) | ||||||||
| M38 | N | 6.05 | C20H19O8 | 387.10737 | 387.10733 | −0.2 | MS2[387]:372(100),357(4) | Methoxylation |
| MS3[372]:357(100),341(0.5),343(0.5) | ||||||||
| P | 6.10 | C20H21O8 | 389.12302 | 389.12207 | −2.6 | MS2[389]:328(100),374(66),345(29),359(28),356(22),373(20) | ||
| MS3[328]:313(100),267(82),312(66),211(64),297(53),284(37) | ||||||||
| M39 | N | 7.30 | C20H19O8 | 387.10737 | 387.10739 | −0.1 | MS2[387]:372(100),357(2) | Methoxylation |
| MS3[372]:357(100),341(1),343(0.6) | ||||||||
| M40 | N | 7.60 | C20H19O8 | 387.10737 | 387.10764 | 0.5 | MS2[387]:372(100),357(5),151(1) | Methoxylation |
| MS3[372]:357(100),341(1),343(0.6) | ||||||||
| M41 | N | 8.76 | C20H19O8 | 387.10737 | 387.10693 | −1.3 | MS2[387]:372(100),388(29),357(9),373(3) | Methoxylation |
| MS3[372]:357(100),341(1),343(0.5) | ||||||||
| P | 8.76 | C20H21O8 | 389.12302 | 389.12198 | −2.8 | MS2[389]:328(100),374(57),390(47),345(28),359(27),355(3) | ||
| MS3[328]:313(100),312(78),267(69),211(48),297(41),296(33),300(28) | ||||||||
| M42 | P | 9.08 | C20H21O8 | 389.12302 | 389.12207 | −2.6 | MS2[389]:374(100),356(75),328(19),390(15) | Methoxylation |
| MS3[374]:356(100),328(2) | ||||||||
| M43 | N | 9.50 | C20H19O8 | 387.10737 | 387.10721 | −0.6 | MS2[387]:372(100),357(2),373(1) | Methoxylation |
| MS3[372]:357(100),343(1),341(0.6) | ||||||||
| P | 9.50 | C20H21O8 | 389.12302 | 389.12207 | −2.6 | MS2[389]:374(100),328(63),373(29),345(20),356(16),359(15) | ||
| MS3[374]:328(100),312(44),358(38),345(27),359(17),373(16),356(16) | ||||||||
| M44 | N | 9.80 | C20H19O8 | 387.10737 | 387.10748 | 0.1 | MS2[387]:372(100),357(3),373(1) | Methoxylation |
| MS3[372]:357(100),341(1),343(0.5) | ||||||||
| M45 | P | 13.75 | C20H21O8 | 389.12302 | 389.1221 | −2.5 | MS2[389]:356(100),328(71),374(69),390(42),389(14),359(14) | Methoxylation |
| MS3[356]:328(100),313(1) | ||||||||
| M46 | P | 15.68 | C20H21O8 | 389.12302 | 389.12225 | −2.1 | MS2[389]:374(100),356(60),328(57),359(16),375(14),345(13),373(13),147(1) | Methoxylation |
| M48 | P | 17.76 | C20H21O8 | 389.12302 | 389.12222 | −2.2 | MS2[389]:374(100),328(52),356(50),359(12),345(12),119(1),211(1) | Methoxylation |
| M49 | P | 19.47 | C20H21O8 | 389.12302 | 389.12241 | −1.7 | MS2[389]:374(100),328(56),356(49),359(12),345(12),139(1),158(1) | Methoxylation |
| M50 | P | 25.00 | C20H21O8 | 389.12302 | 389.12231 | −2.0 | MS2[389]:374(100),328(51),356(50),373(15),359(14),345(12),185(2) | Methoxylation |
| M51 | P | 5.19 | C20H23O8 | 391.13867 | 391.13779 | −2.4 | MS2[391]:207(100),211(66),373(66),175(22),147(7) | Methoxylation + Flavanone Formation |
| MS3[207]:195(100),147(5),119(4),179(3),177(1),192(1) | ||||||||
| M52 | N | 5.61 | C20H21O8 | 389.12412 | 389.12289 | −0.5 | MS2[389]:374(100),359(25),341(8),371(5),165(2),307(2) | Methoxylation + Flavanone Formation |
| MS3[374]:359(100),179(2),165(2),208(1) | ||||||||
| M53 | N | 7.19 | C20H21O8 | 389.12412 | 389.12259 | −1.2 | MS2[389]:179(100),374(73),164(33),373(24),359(14),149(13),121(1) | Methoxylation + Flavanone Formation |
| MS3[179]:164(100),149(1) | ||||||||
| M54 | P | 5.72 | C20H23O8 | 391.13867 | 391.13809 | −1.6 | MS2[391]:221(100),373(49),207(34),197(18),175(10) | Methoxylation + Chalcone Formation |
| MS3[221]:193(100),190(49),206(43),191(41),189(9) | ||||||||
| M55 | N | 6.5 | C20H21O8 | 389.12412 | 389.12259 | −1.2 | MS2[389]:374(100),359(36),390(19),165(3),180(3),389(3) | Methoxylation + Chalcone Formation |
| MS3[374]:359(100),165(3),179(3),208(1) | ||||||||
| M56 | N | 6.96 | C20H21O8 | 389.12412 | 389.12292 | −0.4 | MS2[389]:374(100),359(25),165(3),180(2),373(2),347(1) | Methoxylation + Chalcone Formation |
| MS3[374]:359(100),165(3),180(2) | ||||||||
| P | 6.93 | C20H23O8 | 391.13867 | 391.13785 | −2.2 | MS2[391]:221(100),373(42),197(28),392(10),193(5),190(5),206(2),191(2) | ||
| MS3[221]:193(100),190(48),191(41),206(36),189(15) | ||||||||
| M57 | N | 7.64 | C20H21O8 | 389.12412 | 389.12286 | −0.6 | MS2[389]:374(100),359(78),390(43),389(19),165(10),375(9),360(7),180(6),208(4) | Methoxylation + Chalcone Formation |
| MS3[374]:359(100),165(3),179(3),208(1) | ||||||||
| P | 7.64 | C20H23O8 | 391.13867 | 391.13779 | −2.4 | MS2[391]:221(100),373(18),197(17),193(4),190(3),182(2),206(1),191(1) | ||
| MS3[221]:193(100),190(59),206(47),191(39),189(16),178(5) | ||||||||
| M58 | N | 7.78 | C20H21O8 | 389.12412 | 389.12296 | −0.3 | MS2[389]:374(100),359(29),165(3),180(2),347(1) | Methoxylation + Chalcone Formation |
| MS3[374]:359(100),165(4),180(2),343(1),208(1),221(1) | ||||||||
| M59 | P | 12.97 | C20H23O8 | 391.13867 | 391.13809 | −1.6 | MS2[391]:221(100),197(48),373(12),223(8),349(4) | Methoxylation + Chalcone Formation |
| MS3[221]:193(100),190(50),191(40),206(39),189(13),177(6) | ||||||||
| M60 | N | 13.44 | C20H21O8 | 389.12412 | 389.12305 | −0.1 | MS2[389]:374(100),359(39),390(32),389(7),208(4) | Methoxylation + Chalcone Formation |
| MS3[374]:359(100),208(6),165(3),358(2),180(2),193(1) | ||||||||
| P | 13.44 | C20H23O8 | 391.13867 | 391.13776 | −2.5 | MS2[391]:221(100),197(20),373(19),392(19),373(9) | ||
| MS3[221]:193(100),190(54),205(40),191(40),189(15),178(3) | ||||||||
| M61 | N | 9.87 | C20H21O8 | 389.12412 | 389.12305 | −0.1 | MS2[389]:374(100),359(26),208(3),373(2),280(1) | Loss of Carbonyl + Di-Methoxylation |
| MS3[374]:359(100),208(4),165(3),180(3),193(1) | ||||||||
| M62 | N | 10.34 | C20H21O8 | 389.12412 | 389.12292 | −0.4 | MS2[389]:374(100),359(24),208(2),179(1),165(1) | Loss of Carbonyl + Di-Methoxylation |
| MS3[374]:359(100),208(4),165(3),180(2) | ||||||||
| M63 | N | 11.99 | C20H21O8 | 389.12412 | 389.12314 | 0.1 | MS2[389]:179(100),164(33),374(21),149(14),359(2),121(2),205(2) | Loss of Carbonyl + Di-Methoxylation |
| MS3[179]:164(100),149(2) | ||||||||
| M64 | N | 14.97 | C20H21O8 | 389.12412 | 389.12323 | 0.3 | MS2[389]:343(100),353(16),345(12),313(10),374(10),327(9),179(1),195(1) | Loss of Carbonyl + Di-Methoxylation |
| MS3[343]:325(100),299(20),259(17),287(15),271(11),187(10) | ||||||||
| M65 | N | 16.10 | C20H21O8 | 389.12412 | 389.12418 | 2.7 | MS2[389]:345(100),327(61),343(41),353(27),311(13),151(1) | Loss of Carbonyl + Di-Methoxylation |
| MS3[345]:327(100),311(9),325(7),343(7),317(2) | ||||||||
| M66 | N | 10.06 | C19H19O9 | 391.10342 | 391.1022 | −0.4 | MS2[391]:155(100),375(78),140(20),360(16),376(15),221(11),169(7) | Di-Oxidation + Chalcone Formation |
| MS3[155]:140(100),125(15),123(3),95(3) | ||||||||
| M67 | N | 16.11 | C19H19O9 | 391.10342 | 391.10046 | −4.8 | MS2[391]:345(100),347(44),327(37),392(37),329(33),355(31),301(13),343(7) | Di-Oxidation + Flavanone Formation |
| MS3[345]:327(100),325(28),311(10),259(9),343(8),341(5) | ||||||||
| M68 | N | 16.32 | C19H19O9 | 391.10342 | 391.10040 | −3.0 | MS2[391]:345(100),347(44),327(37),329(33),355(31),343(7) | Di-Oxidation + Flavanone Formation |
| MS3[345]:327(100),325(22),311(9),259(5),285(2),301(2) | ||||||||
| M69 | P | 10.54 | C21H23O8 | 403.13877 | 403.13754 | −2.9 | MS2[403]:373(100),388(55),342(41),370(8),387(6),289(4) | Methylation + Methoxylation |
| MS3[373]:345(100),340(64),343(31),358(25),312(17),181(7) | ||||||||
| M70 | P | 11.50 | C21H23O8 | 403.13877 | 403.13748 | −3.1 | MS2[403]:342(100),388(48),343(28),359(24),370(23),343(5),327(3) | Methylation + Methoxylation |
| MS3[342]:327(100),281(62),309(29),151(18),312(18),195(9) | ||||||||
| M71 | P | 12.47 | C21H23O8 | 403.13877 | 403.13745 | −3.2 | MS2[403]:342(100),388(55),373(29),359(25),387(15),343(6),327(2) | Methylation + Methoxylation |
| MS3[342]:327(100),281(78),309(20),314(16),298(16),151(13),195(3) | ||||||||
| M72 | N | 8.54 | C19H17O11S | 453.04857 | 453.04858 | −0.1 | MS2[453]:373(100),358(3) | Sulfate Conjugation |
| MS3[373]:358(100),343(0.6) | ||||||||
| M73 | N | 7.19 | C20H19O11S | 467.06417 | 467.06430 | −0.1 | MS2[467]:387(100),372(8),388(8),452(1) | Methoxylation + Sulfate Conjugation |
| MS3[387]:372(100),357(1) | ||||||||
| M74 | N | 5.94 | C20H21O11S | 469.08092 | 469.08047 | 1.1 | MS2[469]:389(100),259(26),371(6),341(5),374(5),454(4),179(2) | Methoxylation + Sulfate Conjugation and Flavanone Formation |
| MS3[389]:341(100),374(96),371(76),340(33),326(18) | ||||||||
| M75 | N | 6.50 | C20H21O11S | 469.08092 | 469.07980 | −0.23 | MS2[469]:389(100),470(16),390(14),374(1) | Methoxylation + Sulfate Conjugation and Flavanone Formation |
| MS3[389]:372(100),357(1) | ||||||||
| M76 | N | 9.87 | C20H21O11S | 469.08092 | 469.07996 | 0.1 | MS2[469]:389(100) | Methoxylation + Sulfate Conjugation and Flavanone Formation |
| MS3[389]:374(100),359(24),208(2),180(1) | ||||||||
| M77 | N | 7.05 | C24H23O13 | 519.11332 | 519.11322 | −0.1 | MS2[519]:343(100),175(7),501(5),328(5),329(2) | Loss of CH2 + Glucuronide Conjugation |
| MS3[343]:328(100),284(0.1),313(0.1) | ||||||||
| P | 7.07 | C24H25O13 | 521.12897 | 521.12762 | −2.5 | MS2[521]:345(100),346(14),522(7),330(1) | ||
| MS3[345]:330(100),312(89),284(3),345(2),327(2) | ||||||||
| M78 | N | 7.49 | C25H25O13 | 533.12897 | 533.12878 | −0.3 | MS2[533]:357(100),175(2),342(0.5) | Glucuronide Conjugation |
| MS3[357]:342(100),327(14) | ||||||||
| M79 | N | 4.99 | C25H27O13 | 535.14572 | 535.14508 | 0.8 | MS2[533]:359(100),345(96),175(43),517(22),359(19),212(6),147(6),197(6) | Glucuronide Conjugation + Flavanone Formation |
| M80 | N | 5.75 | C25H25O14 | 549.12387 | 549.12384 | −0.1 | MS2[549]:373(100),358(2),487(0.5),353(0.4) | Oxidation + Glucuronide Conjugation |
| MS3[373]:358(100),343(3),286(0.6),297(0.2) | ||||||||
| P | 5.80 | C25H27O14 | 551.13952 | 551.13818 | −2.4 | MS2[551]:375(100),191(5),375(3),389(1) | ||
| MS3[375]:314(100),360(73),331(29),345(26),359(26),342(21) | ||||||||
| M81 | N | 7.53 | C25H25O14 | 549.12387 | 549.1236 | −0.5 | MS2[549]:373(100),358(2),487(0.5),353(0.4) | Oxidation + Glucuronide Conjugation |
| MS3[373]:358(100),343(0.4),314(0.1) | ||||||||
| M82 | N | 4.90 | C25H27O14 | 551.14062 | 551.14038 | 1.5 | MS2[551]:375(100),361(7),367(6),175(1) | Oxidation + Glucuronide Conjugation and Flavanone Formation |
| MS3[375]:165(100),150(42),360(11),331(4),316(4),343(3) | ||||||||
| P | 4.89 | C25H29O14 | 553.15517 | 553.15369 | 1.8 | MS2[553]:377(100),211(2),359(0.5),207(0.5) | ||
| MS3[377]:359(100),193(77),211(67),181(23),133(16),342(21) | ||||||||
| M83 | N | 6.05 | C26H27O14 | 563.13947 | 563.13898 | −0.9 | MS2[563]:387(100),388(24),372(3),175(1) | Methoxylation + Glucuronide Conjugation |
| MS3[387]:372(100),357(5) | ||||||||
| M84 | N | 6.52 | C26H27O14 | 563.13947 | 563.13855 | −1.7 | MS2[563]:387(100),388(23),372(21),357(4),175(2) | Methoxylation + Glucuronide Conjugation |
| MS3[387]:372(100),357(1) | ||||||||
| M85 | N | 6.73 | C26H27O14 | 563.13947 | 563.13904 | −0.8 | MS2[563]:387(100),388(26),373(13),372(10),175(2) | Methoxylation + Glucuronide Conjugation |
| MS3[387]:372(100),357(12) | ||||||||
| M86 | N | 6.96 | C26H27O14 | 563.13947 | 563.13892 | −1.0 | MS2[563]:387(100),373(7),175(3),372(3) | Methoxylation + Glucuronide Conjugation |
| MS3[387]:372(100),357(10) |
| No | Peak | tR /min | Formula [M + H]+ | Theoretical Mass m/z | Experimental Mass m/z | Error (ppm) | Identification/ Reactions |
|---|---|---|---|---|---|---|---|
| N1 | P | 10.78 | C15H11O6 | 287.05497 | 287.05466 | −1.2 | Loss of 3CH2 + Demethoxylation |
| N2 | P | 8.46 | C16H13O6 | 301.07062 | 301.07025 | −1.3 | Loss of 2CH2 + Demethoxylation |
| N3 | P | 6.98 | C17H15O6 | 315.08627 | 315.08646 | 0.4 | Loss of CH2 + Methoxylation |
| N4 | P | 8.84 | C17H15O6 | 315.08627 | 315.08572 | −1.8 | Loss of CH2 + Methoxylation |
| N5 | P | 12.83 | C17H15O6 | 315.08627 | 315.08624 | −0.2 | Loss of CH2 + Methoxylation |
| N6 | P | 8.78 | C18H19O6 | 331.11761 | 331.11691 | −2.1 | Loss of OCH2 + Flavanone/Chalcone Formation |
| N7 | P | 7.96 | C17H17O7 | 333.09697 | 333.09683 | −0.1 | Loss of 2CH2 + Flavanone/Chalcone Formation |
| N8 | P | 7.38 | C17H13O8 | 345.06042 | 345.06033 | −0.4 | Loss of CH4 and CH2 + Oxidation |
| N9 | P | 10.03 | C19H21O9 | 393.11797 | 393.11673 | −3.2 | Di-Oxidation + Flavanone/Chalcone Formation |
| N10 | N | 7.86 | C17H13O10S | 409.02237 | 409.02164 | −1.8 | Loss of 2CH2 + Sulfate Conjugation |
| N11 | P | 9.38 | C17H15O10S | 411.03802 | 411.03696 | −2.6 | Loss of 2CH2 +Sulfate Conjugation |
| N12 | N | 8.19 | C18H15O10S | 423.03802 | 423.03772 | −0.7 | Loss of CH2 + Sulfate Conjugation |
| N13 | N | 4.77 | C18H17O10S | 425.05472 | 425.05362 | −0.1 | Loss of CH2 + Flavanone/Chalcone Formation and Sulfate Conjugation |
| N14 | P | 5.36 | C19H21O10S | 441.08497 | 441.08429 | −1.5 | Flavanone/Chalcone Formation + Sulfate Conjugation |
| N15 | N | 6.84 | C23H21O13 | 505.09767 | 505.09750 | −0.3 | Loss of 2CH2 + Glucuronide Conjugation |
| N16 | P | 6.81 | C23H23O13 | 507.11332 | 507.11282 | −0.9 | Loss of 2CH2 + Glucuronide Conjugation |
| N17 | P | 7.21 | C24H27O12 | 507.14971 | 507.14813 | −3.1 | Loss of OCH2 + Flavanone/Chalcone Formation and Glucuronide Conjugation |
| N18 | P | 5.57 | C24H23O13 | 519.11332 | 519.11176 | −3.0 | Loss of CH4 + Glucuronide Conjugation |
| N19 | P | 6.74 | C24H25O14 | 537.12387 | 537.12378 | −0.1 | Loss of CH2 + Oxidation and Glucuronide Conjugation |
 | |||||
|---|---|---|---|---|---|
| No | PMFs | Formula | –OH | –OCH3 | Mass Weight |
| P-1 | 5-hydroxy-6,7,3′,4′-tetramethoxyflavone | C19H18O7 | 5 | 6,7,3′,4′ | 358 |
| P-2 | 5-hydroxy-7,3′,4′-trimethoxyflavone | C18H16O6 | 5 | 7,3′,4′ | 328 |
| P-6 | 5-hydroxy-6,7,8,3′,4′-pentamethoxyflavone | C20H20O8 | 5 | 6,7,8,3′,4′ | 388 |
| P-7 | 5-hydroxy-6,7,3′,4′,5′-pentamethoxyflavone | C20H20O8 | 5 | 6,7,3′,4′,5′ | 388 |
| P-9 | 5-hydroxy-6,7,8,3′,4′,5′-hexamethoxyflavone | C21H22O9 | 5 | 6,7,8,3′,4′,5′ | 418 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Mei, X.; Liu, Z.; Li, J.; Zhang, X.; Lang, S.; Dai, L.; Zhang, J. Drug Metabolite Cluster-Based Data-Mining Method for Comprehensive Metabolism Study of 5-hydroxy-6,7,3′,4′-tetramethoxyflavone in Rats. Molecules 2019, 24, 3278. https://doi.org/10.3390/molecules24183278
Wang Y, Mei X, Liu Z, Li J, Zhang X, Lang S, Dai L, Zhang J. Drug Metabolite Cluster-Based Data-Mining Method for Comprehensive Metabolism Study of 5-hydroxy-6,7,3′,4′-tetramethoxyflavone in Rats. Molecules. 2019; 24(18):3278. https://doi.org/10.3390/molecules24183278
Chicago/Turabian StyleWang, Yuqi, Xiaodan Mei, Zihan Liu, Jie Li, Xiaoxin Zhang, Shuang Lang, Long Dai, and Jiayu Zhang. 2019. "Drug Metabolite Cluster-Based Data-Mining Method for Comprehensive Metabolism Study of 5-hydroxy-6,7,3′,4′-tetramethoxyflavone in Rats" Molecules 24, no. 18: 3278. https://doi.org/10.3390/molecules24183278




