Transcriptome and Metabolite Profiling Reveal Novel Insights into Volatile Heterosis in the Tea Plant (Camellia Sinensis)
Abstract
:1. Introduction
2. Results
2.1. The Volatile Content was Significantly Increased in Tea Leaves from the F1 Hybrids
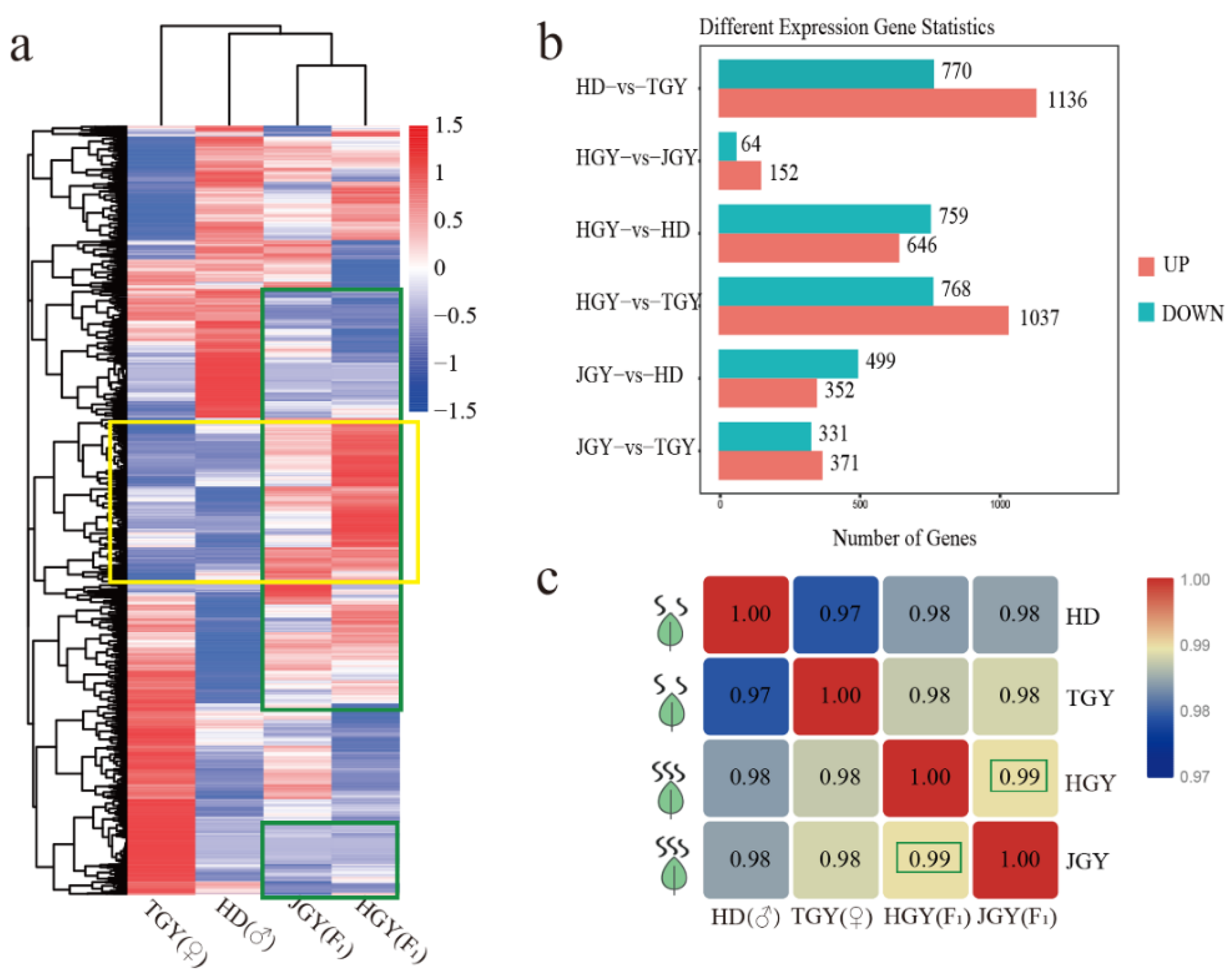
2.2. Higher Gene Expression Similarities were Observed Between the Two F1 Hybrids Than with the Parental Lines
2.3. Expression Level Dominance in the Tea Leaf Transcriptome
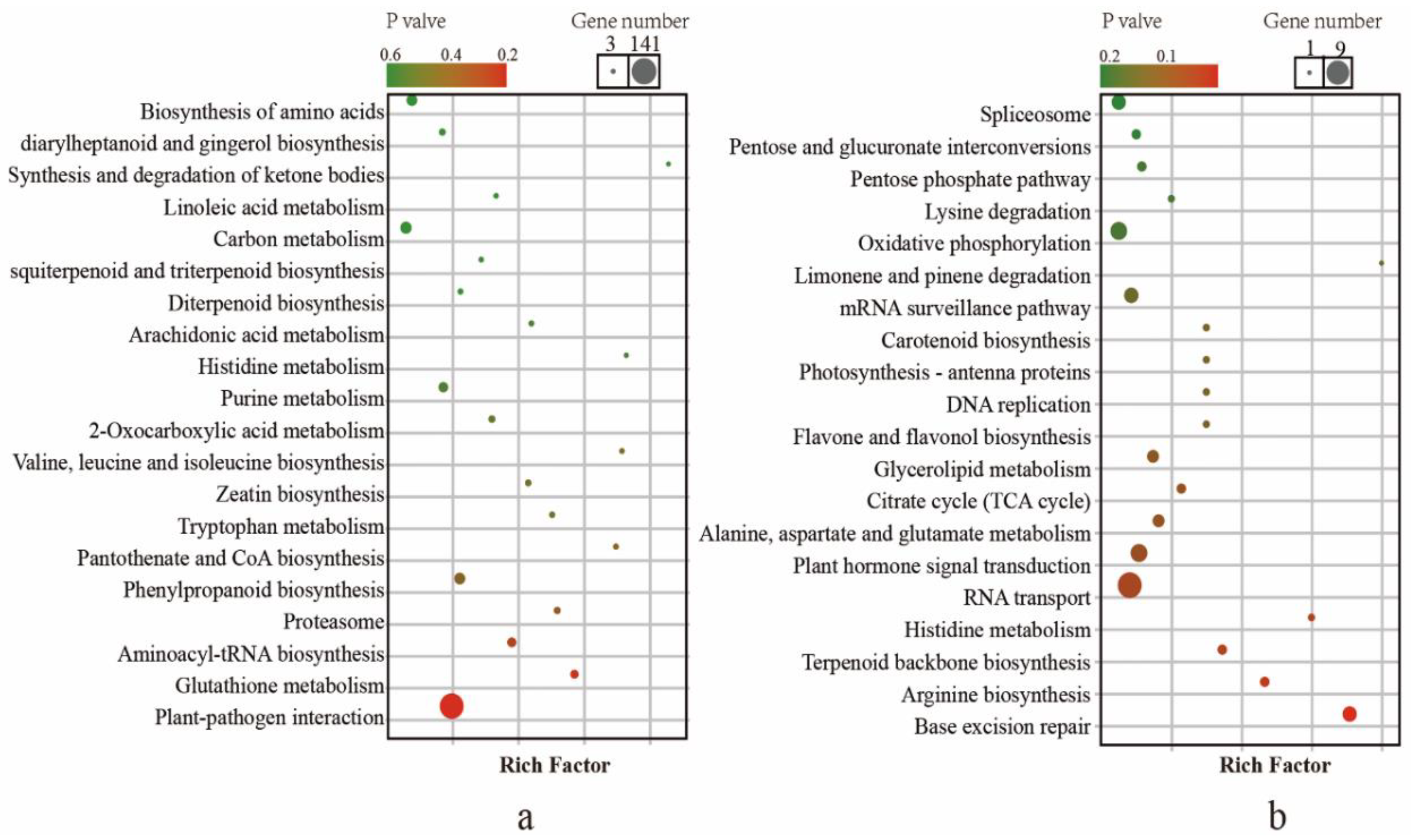
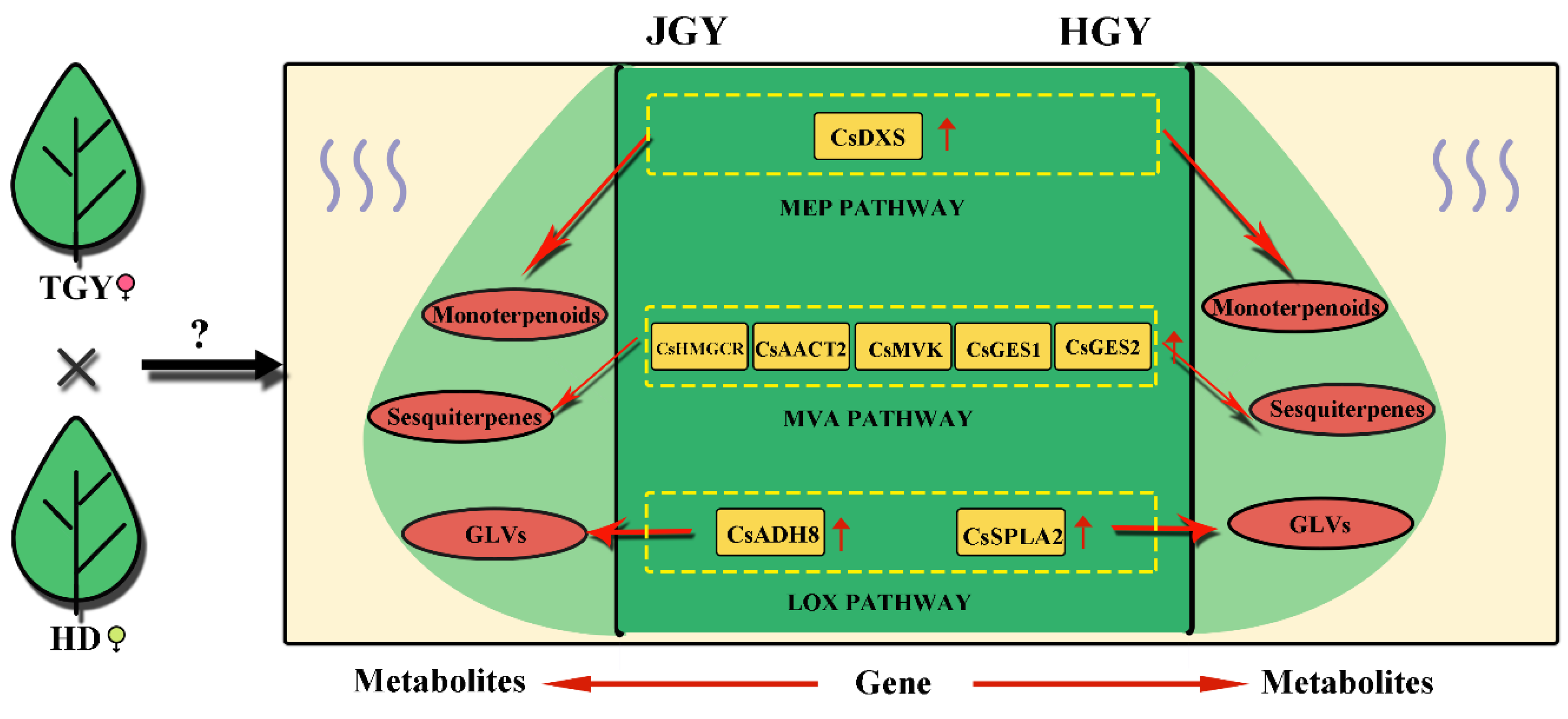
2.4. Analysis of Pathways Related to Tea Aroma
2.5. Identification of TFs Involved in the Regulation of Tea Plant Aroma
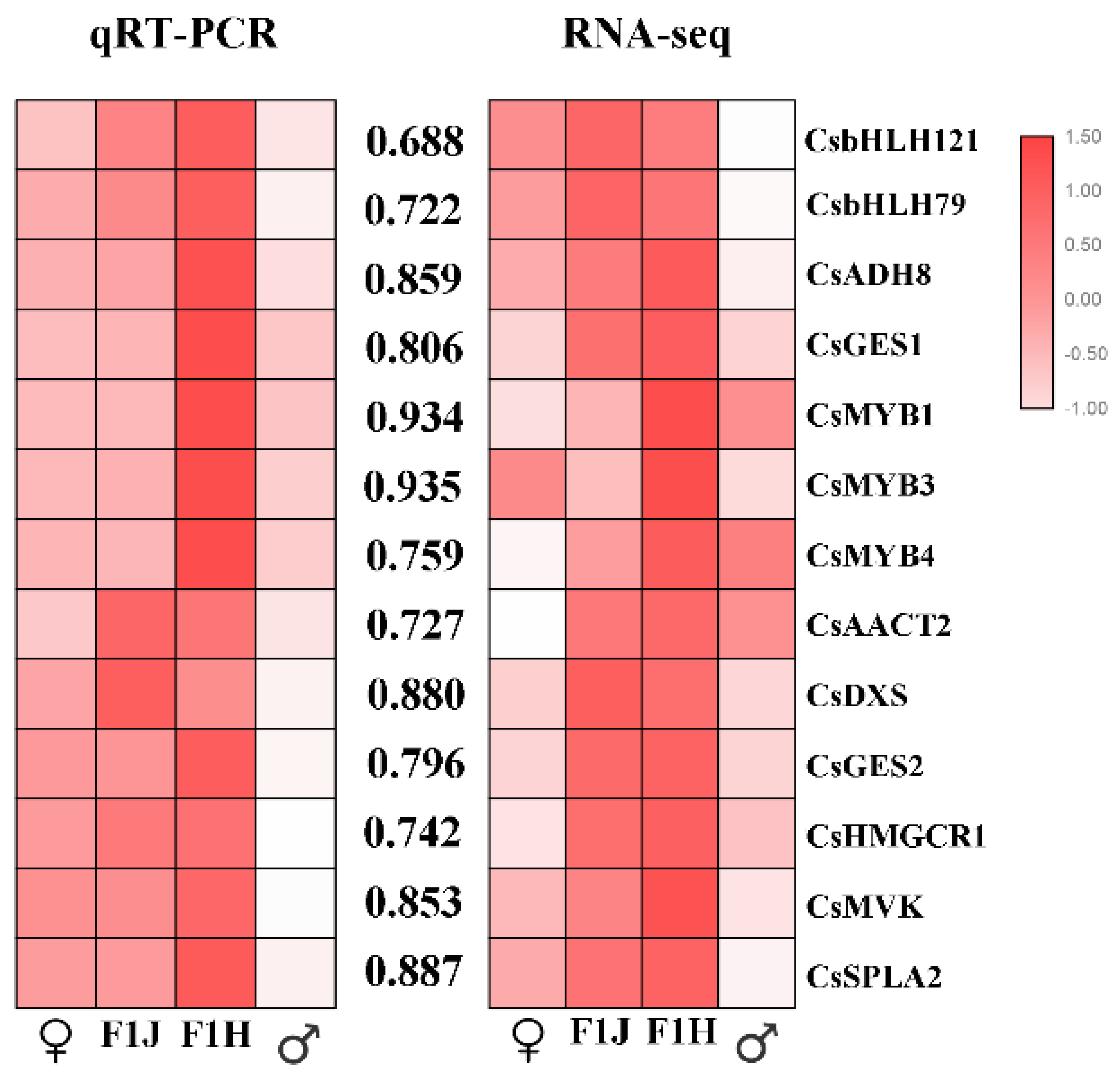
2.6. Gene Expression Validated by qRT-PCR
3. Discussion
3.1. More Meticulous inter-Gene Collaboration May Contribute to Heterosis
3.2. Non-Additive Expression Plays A Dominant Role in The Formation of Tea Heterosis
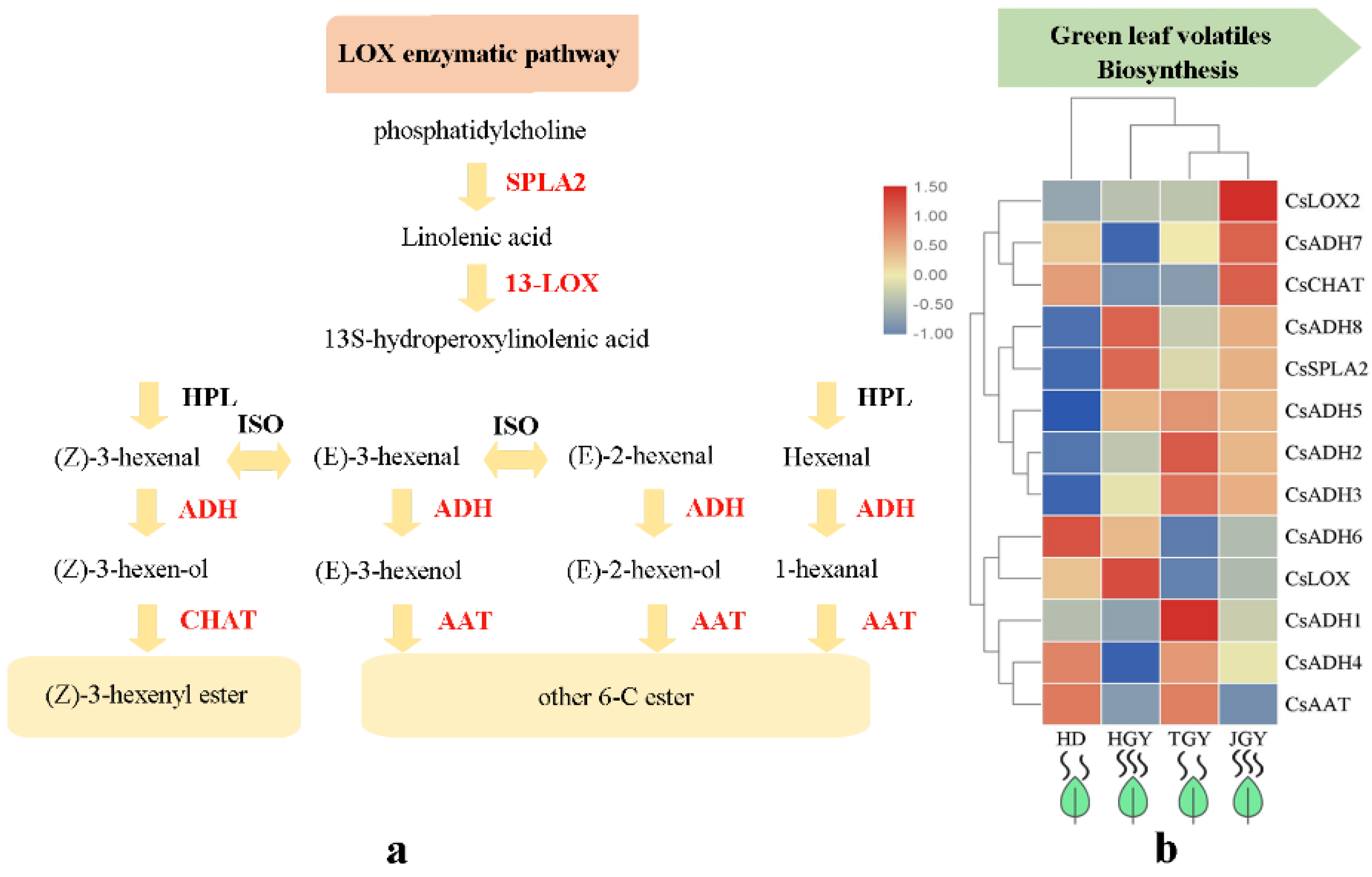
3.3. Genes with Altered Expression Levels May Contribute to Volatile Heterosis
3.4. Transcription Factors (Tfs) Probably Underlie Heterosis
4. Materials and Methods
4.1. Plant Material Preparation
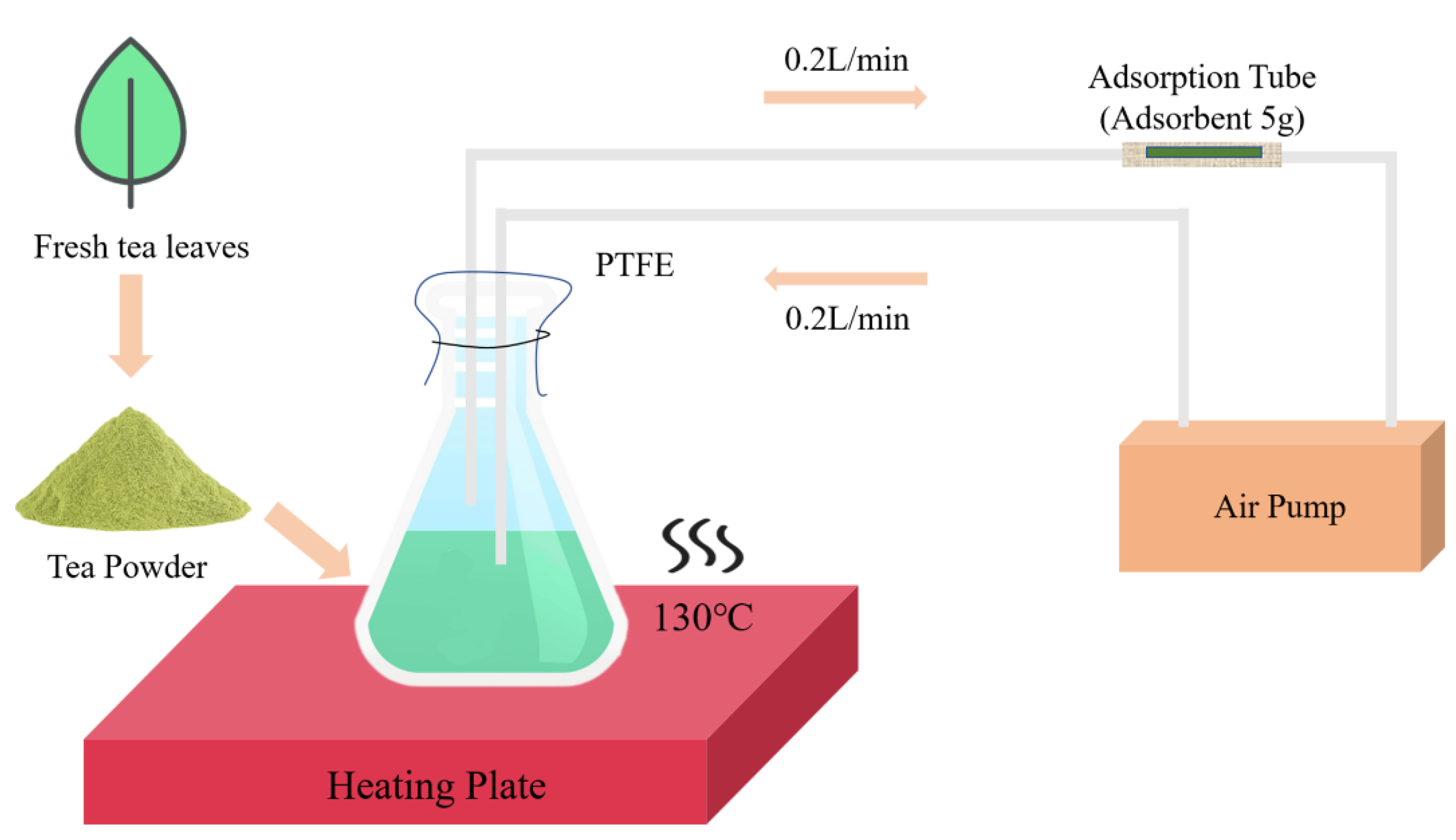
4.2. Thermal Desorption Combined with Gas Chromatography Mass Spectrometry (TD-GC-MS) Analysis of Volatile Compounds in the Leaves
4.3. Transcriptome Sequencing Preparation and Data Analysis
4.4. Gene and Expression Annotation
4.5. Cluster and Trend Analysis
4.6. Validation of the Accuracy of the Transcriptome Data by Quantitative Real-Time PCR Analysis
4.7. Definition of Expression Patterns of Degs Between F1 Hybrids and Parental Lines
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Material
References
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 22, 2105–2112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duvick, D.N. Heterosis: Feeding People and Protecting Natural Resources. Genet. Exploit. Heterosis Crops 1997, 19–29. [Google Scholar]
- Duvick, D.N. Biotechnology in the 1930s: The development ofhybrid maize. Nat. Rev. Genet. 2001, 2, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Frank, H.; Nadine, H. Towards the molecular basis of heterosis. Trends Plant Sci. 2007, 12, 427–432. [Google Scholar]
- Li, Z.K.; Luo, L.J.; Mei, H.W.; Shu, Q.Y.; Tabien, R.; Zhong, D.B.; Ying, C.S.; Stansel, J.W.; Khush, G.S.; Paterson, A.H. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 2001, 158, 1737. [Google Scholar] [PubMed]
- Birchler, J.A.; Hong, Y.; Sivanandan, C. Unraveling the genetic basis of hybrid vigor. Proc. Natl. Acad. Sci. USA 2006, 103, 12957–12958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, P.; Vaidya, E.; Rani, R.; Yadav, N.K.; Singh, B.K.; Rai, P.K.; Singh, D. Recent Perspective of Next Generation Sequencing: Applications in Molecular Plant Biology and Crop Improvement. Proc. Natl. Acad. Sci. India 2018, 88, 435–449. [Google Scholar] [CrossRef]
- Frisch, M.; Thiemann, A.; Fu, J.; Schrag, T.A.; Scholten, S.; Melchinger, A.E. Transcriptome-based distance measures for grouping of germplasm and prediction of hybrid performance in maize. Theor. Appl. Genet. 2010, 120, 441–450. [Google Scholar] [CrossRef]
- Pavani, M.; Sundaram, R.M.; Ramesha, M.S.; Kishor, P.B.K.; Kemparaju, K.B. Prediction of heterosis in rice based on divergence of morphological and molecular markers. J. Genet. 2018, 97, 1263–1279. [Google Scholar] [CrossRef]
- Chen, L.; Bian, J.; Shi, S.; Yu, J.; Khanzada, H.; Wassan, G.M.; Zhu, C.; Luo, X.; Tong, S.; Yang, X.; et al. Genetic analysis for the grain number heterosis of a super-hybrid rice WFYT025 combination using RNA-Seq. Rice 2018, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Tian, M.; Nie, Q.; Li, Z.; Zhang, J.; Liu, Y.; Long, Y.; Wang, Z.; Wang, G.; Liu, R. Transcriptomic analysis reveals overdominance playing a critical role in nicotine heterosis in Nicotiana tabacum L. BMC Plant Biol. 2018, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, L.M.; Greaves, I.K.; Zhu, A.; Dennis, E.S.; Peacock, W.J. PIF4-controlled auxin pathway contributes to hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E3555–E3562. [Google Scholar] [CrossRef] [PubMed]
- Groszmann, M.; Gonzalez-Bayon, R.; Lyons, R.L.; Greaves, I.K.; Kazan, K.; Peacock, W.J.; Dennis, E.S. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. USA 2015, 112, E6397–E6406. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Baldermann, S.; Watanabe, N. Recent studies of the volatile compounds in tea. Food Res. Int. 2013, 53, 585–599. [Google Scholar] [CrossRef]
- Zeng, L.; Watanabe, N.; Yang, Z. Understanding the biosyntheses and stress response mechanisms of aroma compounds in tea (Camellia sinensis) to safely and effectively improve tea aroma. Crit. Rev. Food Sci. Nutr. 2018, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Christian, S.; Peter, S. Characterization of the key aroma compounds in the beverage prepared from Darjeeling black tea: Quantitative differences between tea leaves and infusion. J. Agric. Food Chem. 2006, 54, 916–924. [Google Scholar]
- Hu, C.J.; LI, Da.; M, Y.X.; Zhang, W.; Lin, C.; Zheng, X.Q.; Liang, Y.R.; Lu, J.L. Formation mechanism of the oolong tea characteristic aroma during bruising and withering treatment. Food Chem. 2018, 269, 202–211. [Google Scholar] [CrossRef]
- Vranova, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Zhang, M.; Li, K.; Zhang, C.; Gai, J.; Yu, D. Identification and characterization of class 1 DXS gene encoding 1-deoxy-D-xylulose-5-phosphate synthase, the first committed enzyme of the MEP pathway from soybean. Mol. Biol. Rep. 2009, 36, 879–887. [Google Scholar] [CrossRef]
- Jin, H.; Song, Z.; Nikolau, B.J. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. Plant J. 2012, 70, 1015–1032. [Google Scholar] [CrossRef]
- Zhang, B.; Shen, J.Y.; Wei, W.W.; Xi, W.P.; Xu, C.J.; Ferguson, I.; Chen, K. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J. Agric. Food Chem. 2010, 58, 6157–6165. [Google Scholar] [CrossRef] [PubMed]
- Butt, M.S.; Sultan, M.T. Green Tea: Nature’s Defense against Malignancies. Crit. Rev. Food Sci. Nutr. 2009, 49, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Sabour, M.; Simmonds, J.; Setterfield, G. Variation in Nicotine Content of Cultured Cell Lines of Nicotiana Species and their Somatic and Sexual Hybrids. Plant Breed. 2010, 97, 324–333. [Google Scholar] [CrossRef]
- Marnik, V.; Fred, V.E.; Paul, V.H.; Martin, K.; Marc, Z. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 2005, 171, 1267. [Google Scholar]
- Ryo, F.; Taylor, J.M.; Sachiko, S.; James, P.W.; Dennis, E.S. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. USA 2012, 109, 7109–7114. [Google Scholar]
- Andorf, S.; Selbig, J.; Altmann, T.; Poos, K.; Witucka-Wall, H.; Repsilber, D. Enriched partial correlations in genome-wide gene expression profiles of hybrids (A. thaliana): A systems biological approach towards the molecular basis of heterosis. Theor. Appl. Genet. 2010, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Chen, W.; Song, S.; Wang, W.; Hu, S.; Yu, J. Transcriptomic profiling of mature embryo from an elite super-hybrid rice LYP9 and its parental lines. BMC Plant Boil. 2008, 8, 114. [Google Scholar] [CrossRef]
- Swanson-Wagner, R.A.; Jia, Y.; DeCook, R.; Borsuk, L.A.; Nettleton, D.; Schnable, P.S. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. 2006, 103, 6805–6810. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Yang, X.; Crasta, O.; Zinselmeier, C.; Smith, O.S.; Bowen, B. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 2006, 113, 831–845. [Google Scholar] [CrossRef]
- Springer, N.M.; Stupar, R.M. Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Res. 2007, 17, 264–275. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.; Riley-Berger, R.; Harshman, L.; Kopp, A.; Vacha, S.; Nuzhdin, S.; Wayne, M. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 2004, 167, 1791–1799. [Google Scholar] [CrossRef]
- Uzarowska, A.; Keller, B.; Piepho, H.P.; Schwarz, G.; Ingvardsen, C.; Wenzel, G.; Lubberstedt, T. Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol. Biol. 2007, 63, 21–34. [Google Scholar] [CrossRef]
- Li, A.; Liu, D.; Wu, J.; Zhao, X.; Hao, M.; Geng, S.; Yan, J.; Jiang, X.; Zhang, L.; Wu, J.; et al. mRNA and Small RNA Transcriptomes Reveal Insights into Dynamic Homoeolog Regulation of Allopolyploid Heterosis in Nascent Hexaploid Wheat[W][OPEN]. Plant Cell 2014, 26, 1878–1900. [Google Scholar] [CrossRef]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef]
- Birchler, J.A.; Riddle, N.C.; Auger, D.L.; Veitia, R.A. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005, 21, 219–226. [Google Scholar] [CrossRef]
- Lange, B.M.; Wildung, M.R.; McCaskill, D.; Croteau, R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 2100–2104. [Google Scholar] [CrossRef] [Green Version]
- Estevez, J.M.; Romero, C.K.H.; Jimenez, L.F.; Kuzuyama, T.S.H.; Kamiya, Y.; Leon, P.; Cantero, A. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000, 124, 95–103. [Google Scholar] [CrossRef]
- Morris, W.L.; Ducreux, L.J.M.; Hedden, P.; Millam, S.; Taylor, M.A. Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: implications for the control of the tuber life cycle. J. Exp. Bot. 2006, 57, 3007–3018. [Google Scholar] [CrossRef] [Green Version]
- Enfissi, E.M.A.; Fraser, P.D.; Lois, L.M.; Boronat, A.; Schuch, W.; Bramley, P.M. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 2010, 3, 17–27. [Google Scholar] [CrossRef]
- Rodríguez-Concepción, M. Early Steps in Isoprenoid Biosynthesis: Multilevel Regulation of the Supply of Common Precursors in Plant Cells. Phytochem. Rev. 2006, 5, 1–15. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, O.R.; Oh, J.Y.; Jang, M.G.; Yang, D.C. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014, 165, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Lluch, M.A.; Masferrer, A.; Arro, M.; Boronat, A.; Ferrer, A. Molecular cloning and expression analysis of the mevalonate kinase gene from Arabidopsis thaliana. Plant Mol. Biol. 2000, 42, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Dolferus, R.; Jacobs, M.; Peacock, W.J.; Dennis, E.S. Differential Interactions of Promoter Elements in Stress Responses of the Arabidopsis Adh Gene. Plant Physiol. 1994, 105, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; He, H.; Chen, L.B.; Li, L.; Liang, M.Z.; Wang, X.F.; Liu, X.G.; He, G.M.; Chen, R.S.; Ma, L.G.; et al. A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol. Plant 2008, 1, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed]
- Bedon, F.; Bomal, C.; Caron, S.; Levasseur, C.; Boyle, B.; Mansfield, S.D.; Schmidt, A.; Gershenzon, J.; Grimapettenati, J.; Séguin, A. Subgroup 4 R2R3-MYBs in conifer trees: Gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 2010, 61, 3847–3864. [Google Scholar] [CrossRef] [PubMed]
- Skibbe, M.; Qu, N.; Galis, I.; Baldwin, I.T. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 2008, 20, 1984–2000. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, P.C.; Liu, P.P.; Song, X.W.; Guo, F.; Li, Y.Y.; Ni, D.J.; Jiang, C.J. Novel insight into the role of withering process in characteristic flavor formation of teas using transcriptome analysis and metabolite profiling. Food Chem. 2019, 272, 313–322. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, 201719622. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |




| RT | CAS | RI | MS | Compounds | HGY | JGY | TGY | HD |
|---|---|---|---|---|---|---|---|---|
| 5.78 | 66-25-1 | 806 | 96 | Hexanal | 56.1 ± 12.86a | 91.4 ± 17.77b | 43.92 ± 11.85a | 42.33 ± 5.75a |
| 7.36 | 6728-26-3 | 814 | 95 | (E)-2-Hexenal | 72.5 ± 21.88a | 225.95 ± 40.01b | 127.13 ± 11.51c | 60.23 ± 8.07a |
| 7.47 | 928-97-2 | 868 | 96 | (E)-3-Hexen-1-ol | 92.84 ± 30.95b | 80.86 ± 7.94b | 77.22 ± 3.49b | 84.44 ± 22.07a |
| 7.76 | 928-94-9 | 868 | 97 | (Z)-2-Hexen-1-ol | 10.66 ± 1a | 13.78 ± 1.61b | 5.83 ± 0.61c | 3.28 ± 0.59d |
| 7.89 | 111-27-3 | 860 | 92 | 1-Hexanol | 84.97 ± 14.18a | 69.65 ± 8.29a | 33.79 ± 12.23b | 27.11 ± 9.48b |
| 12.41 | 3681-71-8 | 992 | 96 | (Z)-3-Hexen-1-ol acetate | 116.31 ± 32.14b | 102.98 ± 22.15b | 115.78 ± 19.19b | 66.79 ± 2.6a |
| Sum | (GLVs) | 432.93 ± 79.04bc | 584.62 ± 91.38c | 400.67 ± 66.53b | 284.18 ± 40.92a | |||
| 14.73 | 5989-33-3 | 1529 | 92 | Linalool oxide 1 | 289.92 ± 92.76b | 284.78 ± 25.62b | 115.73 ± 23.83a | 190.75 ± 4.42b |
| 15.32 | 34995-77-2 | 1529 | 95 | Linalool oxide 2 | 509.3 ± 98.74a | 287.01 ± 20.36a | 241.48 ± 69.98a | 116.86 ± 59.02a |
| 15.73 | 78-70-6 | 1082 | 94 | Linalool | 387.86 ± 66.16a | 447.12 ± 51.29a | 222.01 ± 73.24b | 310.64 ± 42.1b |
| 19.42 | 432-25-7 | 1204 | 90 | β-Cyclocitral | 11.72 ± 2.29a | 4.15 ± 0.79b | 3.55 ± 1.06b | 2.34 ± 0.83b |
| 19.73 | 106-26-3 | 1228 | 90 | cis-Citral | 8.83 ± 3.18b | 20.42 ± 2.65a | 8.31 ± 0.75b | 2.97 ± 0.23c |
| 19.58 | 106-25-2 | 1228 | 92 | (Z)-Geraniol | 4.88 ± 1.22b | 6.75 ± 1.07b | 7.69 ± 1.71b | 32.9 ± 6.66a |
| 20.67 | 106-24-1 | 1228 | 93 | Geraniol | 288.28 ± 29.56b | 505.67 ± 42.21a | 164.19 ± 26c | 257.69 ± 58.04b |
| Sum | (monoterpene) | 1500.79 ± 396.8a | 1555.90 ± 143.99a | 766.43 ± 117.94b | 916.7 ± 73.39b | |||
| 24.63 | 23986-74-5 | 1515 | 85 | Germacrene D | 5.49 ± 1.01a | 1.27 ± 0.08b | 0.34 ± 0.09b | 0.47 ± 0.05b |
| 26.14 | 15423-57-1 | 1344 | 84 | Germacrene B | 6.14 ± 1.76a | 1.47 ± 0.17b | 1.39 ± 0.33b | 1.02 ± 0.15b |
| 27.14 | 502-61-4 | 1458 | 82 | α-Farnesene | 0a | 7.5 ± 0.64b | 0a | 0a |
| 27.49 | 16728-99-7 | 1344 | 86 | Cubenene | 10.21 ± 1.22a | 0c | 6.4 ± 1.61b | 0c |
| 27.68 | 142-50-7 | 1564 | 88 | Nerolidol 2 | 20.74 ± 3.78a | 14.74 ± 3.81a | 14.15 ± 1.77a | 14.69 ± 1.23a |
| Sum | (sesquiterpene) | 42.58 ± 6.77c | 24.98 ± 4.19b | 22.28 ± 2.58b | 16.18 ± 1.18a |
| F1 | Compound | Mid-Parent Heterosis Value | Over High-Parent Heterosis Value |
|---|---|---|---|
| HGY | Green leaf volatiles | 26.4% | 8.0% |
| monoterpene | 78.3% | 63.7% | |
| sesquiterpene | 121.4% | 47.7% | |
| JGY | Green leaf volatiles | 70.1% | 45.9% |
| monoterpene | 84.9% | 69.7% | |
| sesquiterpene | 65.9% | 12.1% |
| Sample | Raw Reads | Clean Reads | Clean Bases | Error (%) | Q20 (%) | Q30 (%) | Gc (%) |
|---|---|---|---|---|---|---|---|
| HD_1 | 23334614 | 22395426 | 3.36G | 0.01% | 99.15% | 97.36% | 45.76% |
| HD_2 | 23333820 | 22402952 | 3.36G | 0.01% | 99.10% | 97.25% | 45.67% |
| HD_3 | 23332984 | 22387946 | 3.36G | 0.01% | 97.54% | 94.10% | 45.83% |
| HGY_1 | 23333348 | 22889486 | 3.43G | 0.01% | 99.09% | 97.24% | 45.39% |
| HGY_2 | 23334211 | 22772393 | 3.43G | 0.01% | 99.12% | 97.33% | 45.37% |
| HGY_3 | 23334388 | 22809487 | 3.43G | 0.01% | 99.10% | 97.27% | 45.57% |
| JGY_1 | 23332300 | 22881487 | 3.42G | 0.01% | 99.09% | 97.24% | 45.25% |
| JGY_2 | 23332329 | 22766832 | 3.42G | 0.01% | 99.10% | 97.24% | 45.97% |
| JGY_3 | 23332293 | 22850808 | 3.42G | 0.01% | 99.12% | 97.30% | 45.34% |
| TGY_1 | 23333500 | 22974617 | 3.45G | 0.01% | 99.14% | 97.35% | 45.03% |
| TGY_2 | 23333469 | 22667735 | 3.45G | 0.01% | 99.13% | 97.34% | 45.48% |
| TGY_3 | 23333552 | 22818189 | 3.45G | 0.01% | 99.12% | 97.32% | 45.25% |
| Total | 280000808 | 272617358 | 40.98G | ||||
| GO ID | GO Term | Gene Ratio | p-value | FDR |
|---|---|---|---|---|
| Higher parent dominance | ||||
| GO:0006528 | asparagine metabolic process | 6 | 0.000721 | 0.516173 |
| GO:0006529 | asparagine biosynthetic process | 6 | 0.000721 | 0.516173 |
| GO:0044237 | cellular metabolic process | 436 | 0.000934 | 0.516173 |
| GO:0010026 | trichome differentiation | 4 | 0.002543 | 0.702621 |
| GO:0010090 | trichome morphogenesis | 4 | 0.002543 | 0.702621 |
| GO:0090558 | plant epidermis development | 7 | 0.003961 | 0.74574 |
| GO:0009888 | tissue development | 13 | 0.00612 | 0.74574 |
| GO:0006796 | phosphate-containing compound metabolic process | 162 | 0.00683 | 0.74574 |
| GO:0006793 | phosphorus metabolic process | 162 | 0.007318 | 0.74574 |
| GO:0000904 | cell morphogenesis involved in differentiation | 5 | 0.007927 | 0.74574 |
| GO:0090626 | plant epidermis morphogenesis | 5 | 0.007927 | 0.74574 |
| GO:0050896 | response to stimulus | 134 | 0.010821 | 0.74574 |
| GO:0019538 | protein metabolic process | 199 | 0.011409 | 0.74574 |
| GO:0006952 | defense response | 30 | 0.018033 | 0.74574 |
| GO:0042742 | defense response to bacterium | 10 | 0.019967 | 0.74574 |
| GO:0009690 | cytokinin metabolic process | 4 | 0.025745 | 0.74574 |
| GO:0009617 | response to bacterium | 10 | 0.027516 | 0.74574 |
| GO:0045595 | regulation of cell differentiation | 3 | 0.037732 | 0.74574 |
| GO:1901566 | organonitrogen compound biosynthetic process | 89 | 0.039279 | 0.74574 |
| GO:0030154 | cell differentiation | 10 | 0.04827 | 0.74574 |
| Up regulated overdominance | ||||
| GO:0008299 | isoprenoid biosynthetic process | 8 | 0.003323 | 0.603651 |
| GO:0016114 | terpenoid biosynthetic process | 6 | 0.010173 | 0.626311 |
| GO:0051321 | meiotic cell cycle | 5 | 0.014234 | 0.626311 |
| GO:0009658 | chloroplast organization | 4 | 0.01434 | 0.626311 |
| GO:0044786 | cell cycle DNA replication | 2 | 0.020165 | 0.626311 |
| GO:0044270 | cellular nitrogen compound catabolic process | 6 | 0.031197 | 0.626311 |
| GO:0009648 | photoperiodism | 2 | 0.038089 | 0.626311 |
| GO:1903046 | meiotic cell cycle process | 4 | 0.040315 | 0.626311 |
| GO:0008610 | lipid biosynthetic process | 12 | 0.040691 | 0.626311 |
| GO:0006721 | terpenoid metabolic process | 6 | 0.041973 | 0.626311 |
| GO:0034004 | germacradienol synthase activity | 3 | 0.01149 | 0.522775 |
| GO:0052577 | germacrene-D synthase activity | 3 | 0.01149 | 0.522775 |
| GO:0010334 | sesquiterpene synthase activity | 3 | 0.01879 | 0.527904 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Wang, P.; Chen, X.; Sun, Y.; Yue, C.; Ye, N. Transcriptome and Metabolite Profiling Reveal Novel Insights into Volatile Heterosis in the Tea Plant (Camellia Sinensis). Molecules 2019, 24, 3380. https://doi.org/10.3390/molecules24183380
Zheng Y, Wang P, Chen X, Sun Y, Yue C, Ye N. Transcriptome and Metabolite Profiling Reveal Novel Insights into Volatile Heterosis in the Tea Plant (Camellia Sinensis). Molecules. 2019; 24(18):3380. https://doi.org/10.3390/molecules24183380
Chicago/Turabian StyleZheng, Yucheng, Pengjie Wang, Xuejin Chen, Yun Sun, Chuan Yue, and Naixing Ye. 2019. "Transcriptome and Metabolite Profiling Reveal Novel Insights into Volatile Heterosis in the Tea Plant (Camellia Sinensis)" Molecules 24, no. 18: 3380. https://doi.org/10.3390/molecules24183380
APA StyleZheng, Y., Wang, P., Chen, X., Sun, Y., Yue, C., & Ye, N. (2019). Transcriptome and Metabolite Profiling Reveal Novel Insights into Volatile Heterosis in the Tea Plant (Camellia Sinensis). Molecules, 24(18), 3380. https://doi.org/10.3390/molecules24183380





