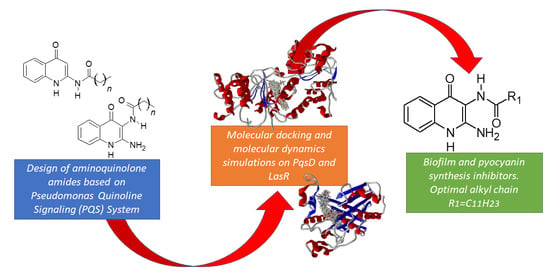
Synthesis, In Silico, and In Vitro Evaluation of Long Chain Alkyl Amides from 2-Amino-4-Quinolone Derivatives as Biofilm Inhibitors
Abstract
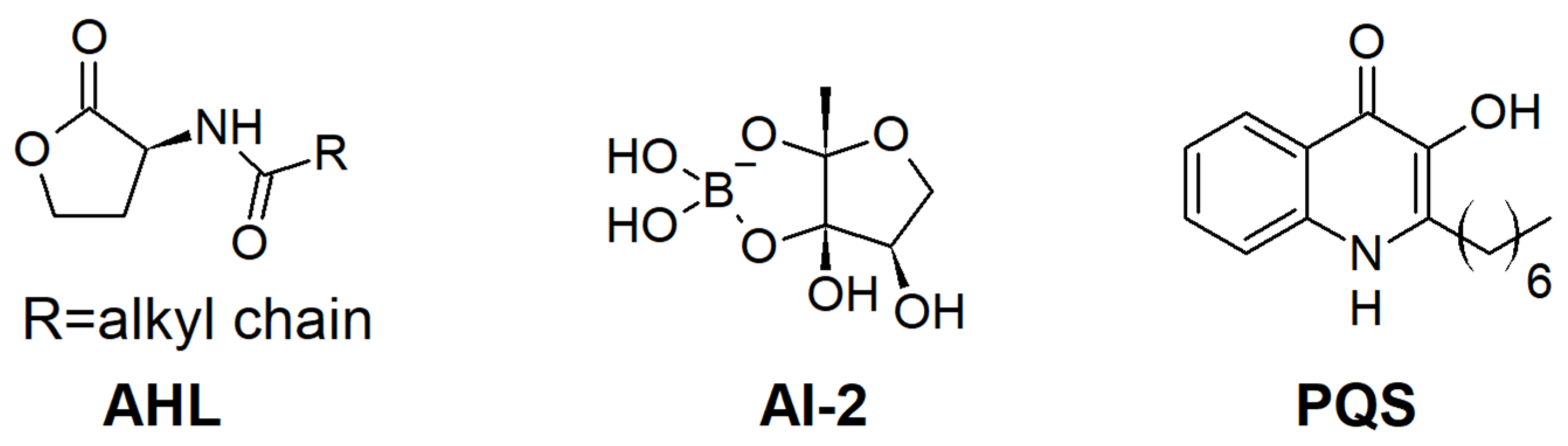
:1. Introduction
2. Results
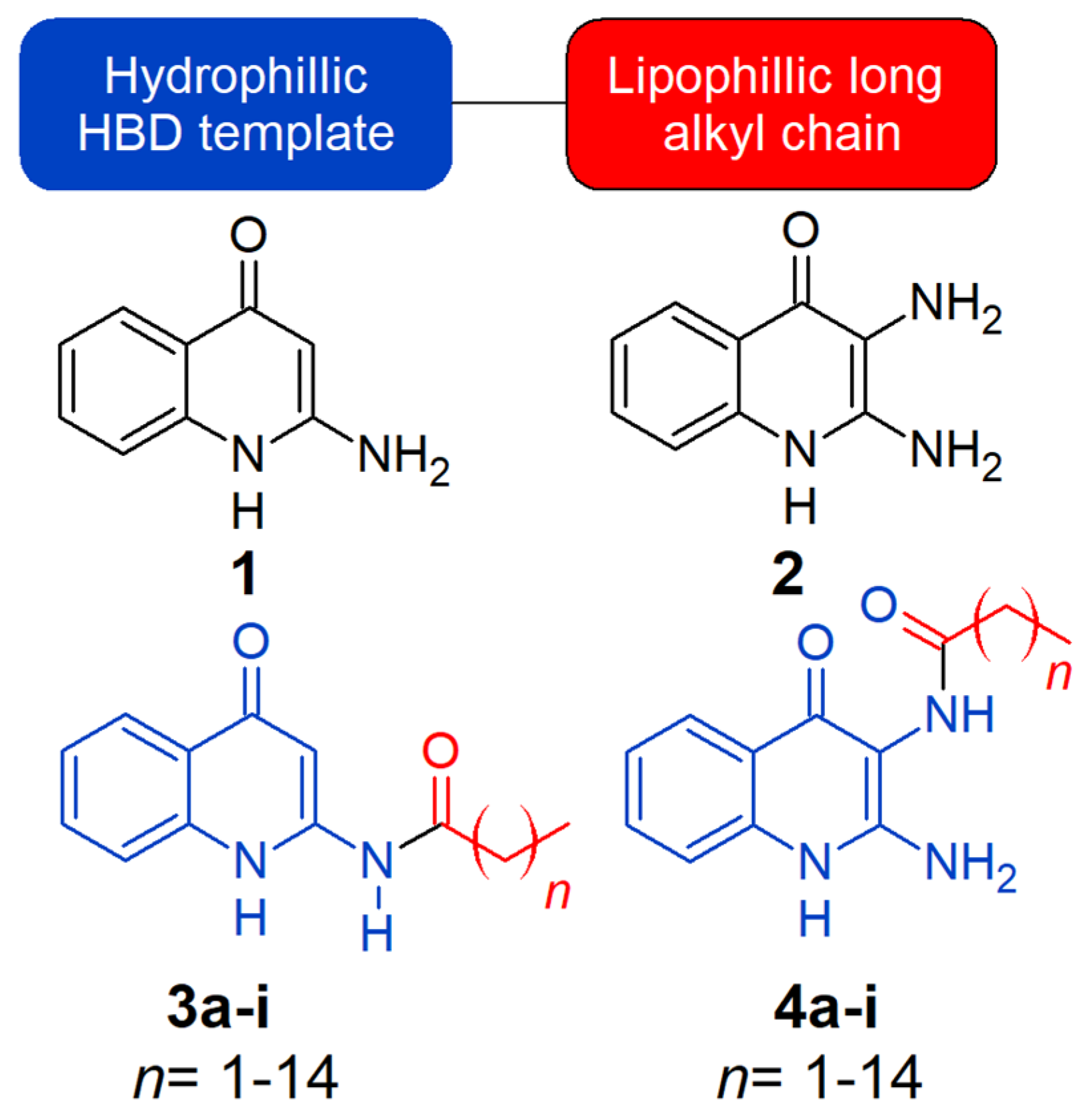
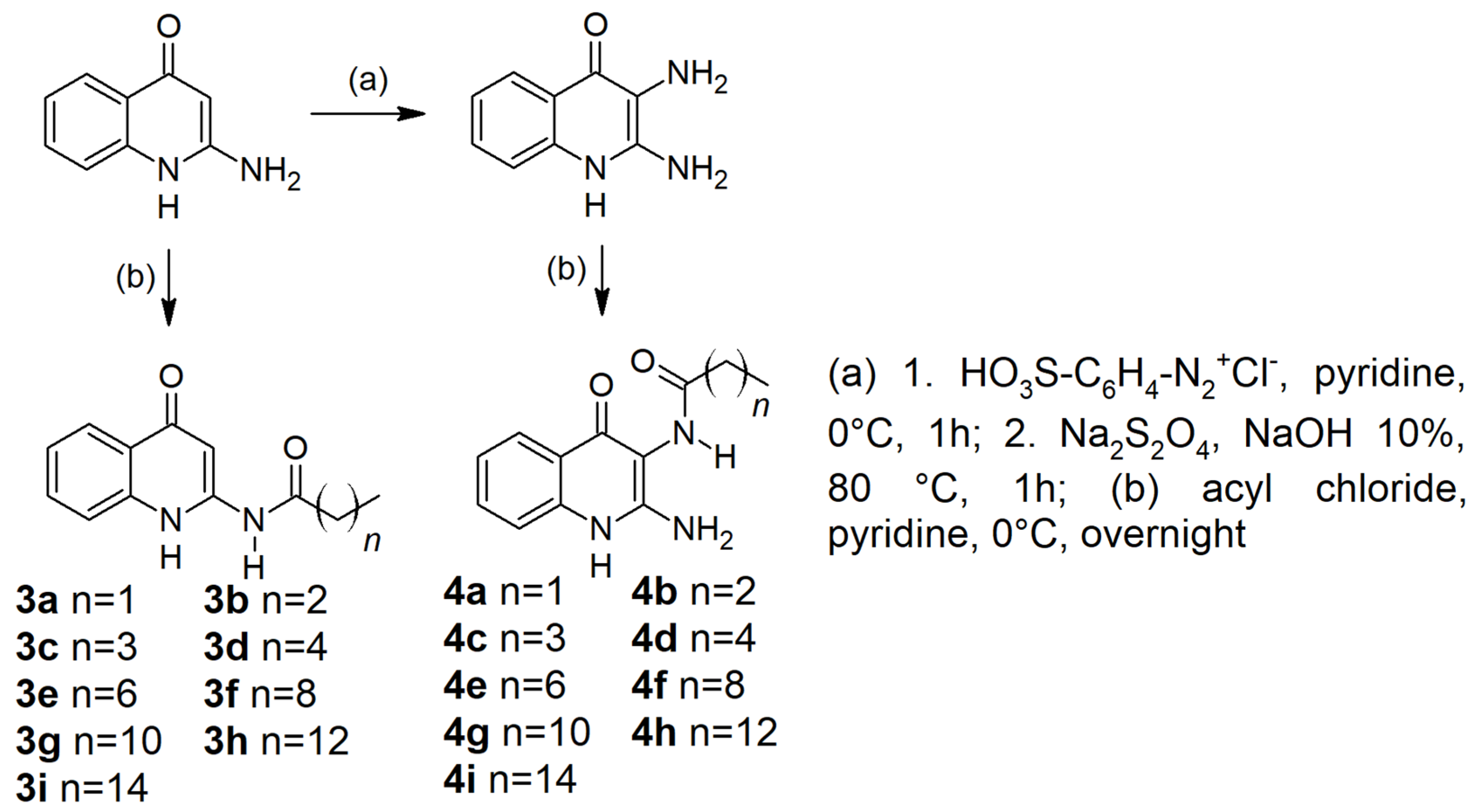
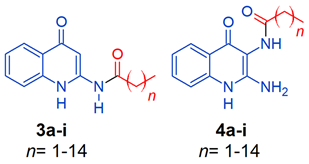
2.1. Chemistry
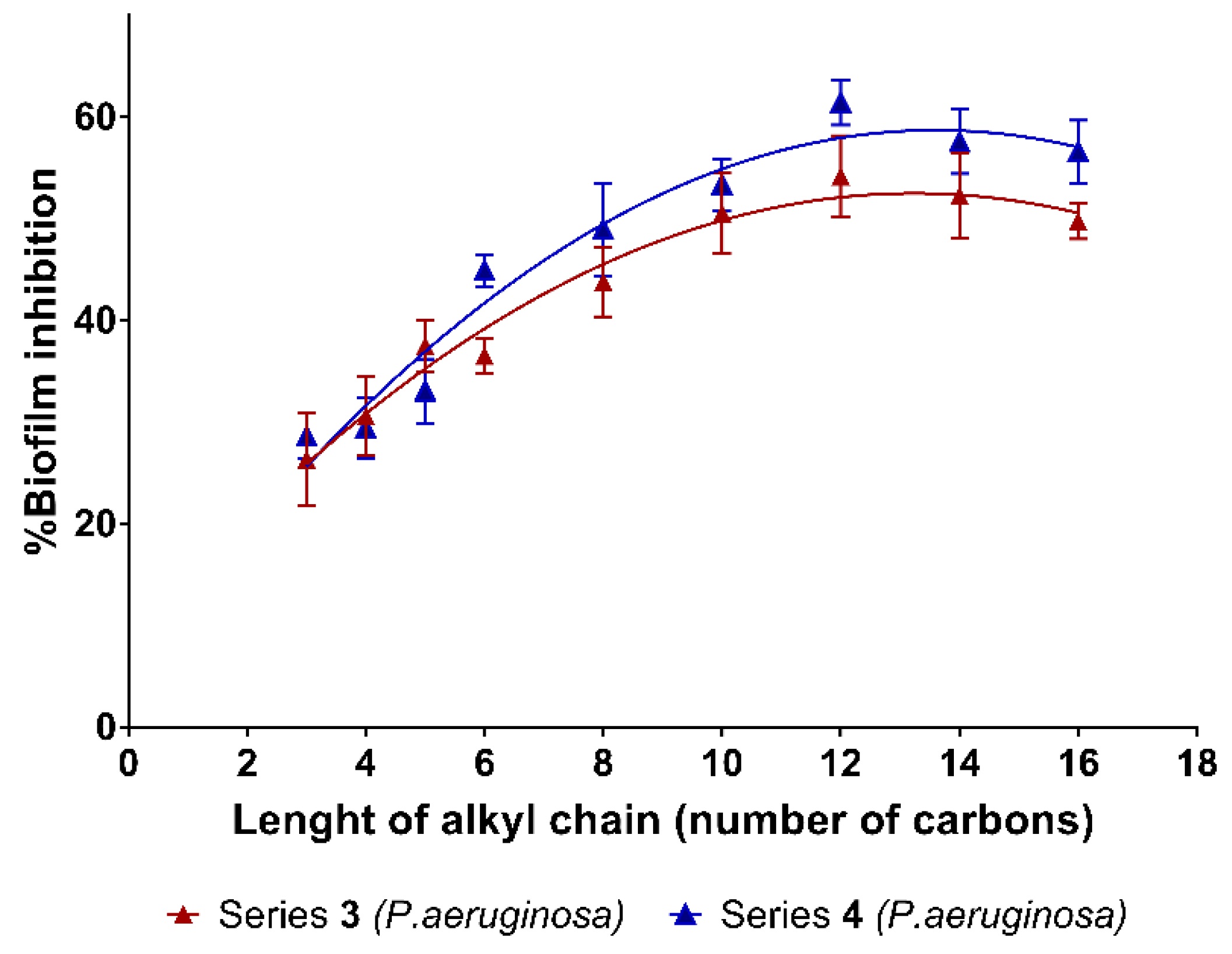
2.2. Biofilm Inhibition
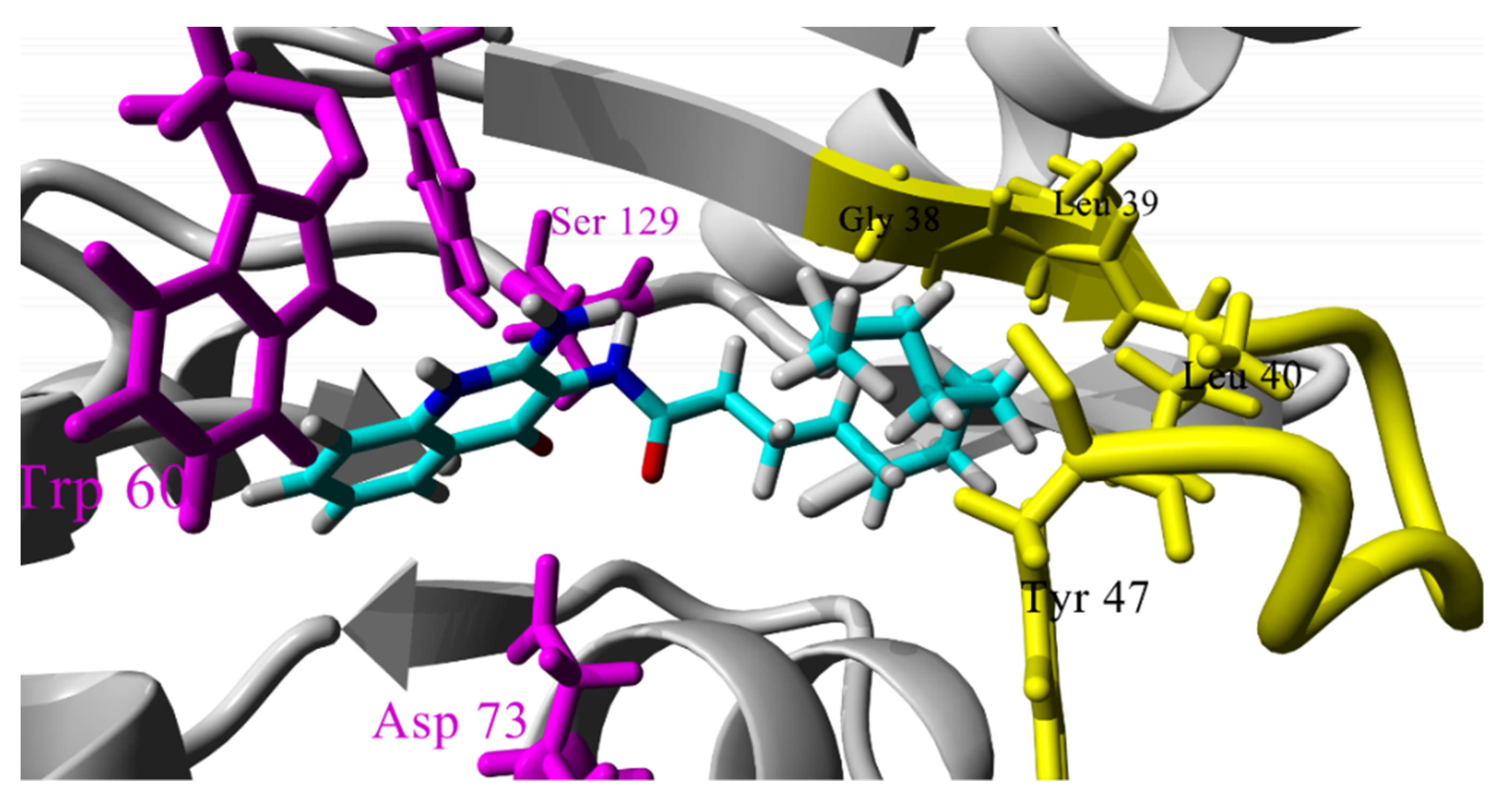
2.3. Molecular Docking Studies
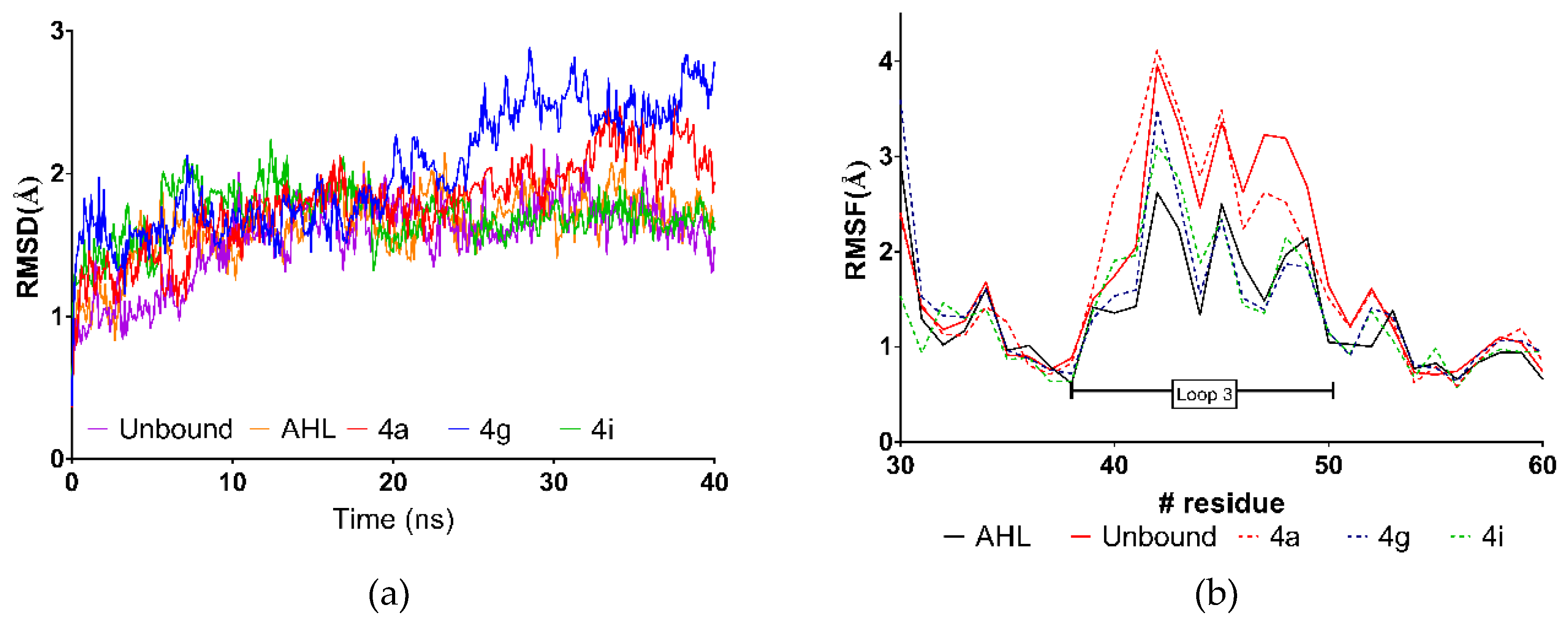
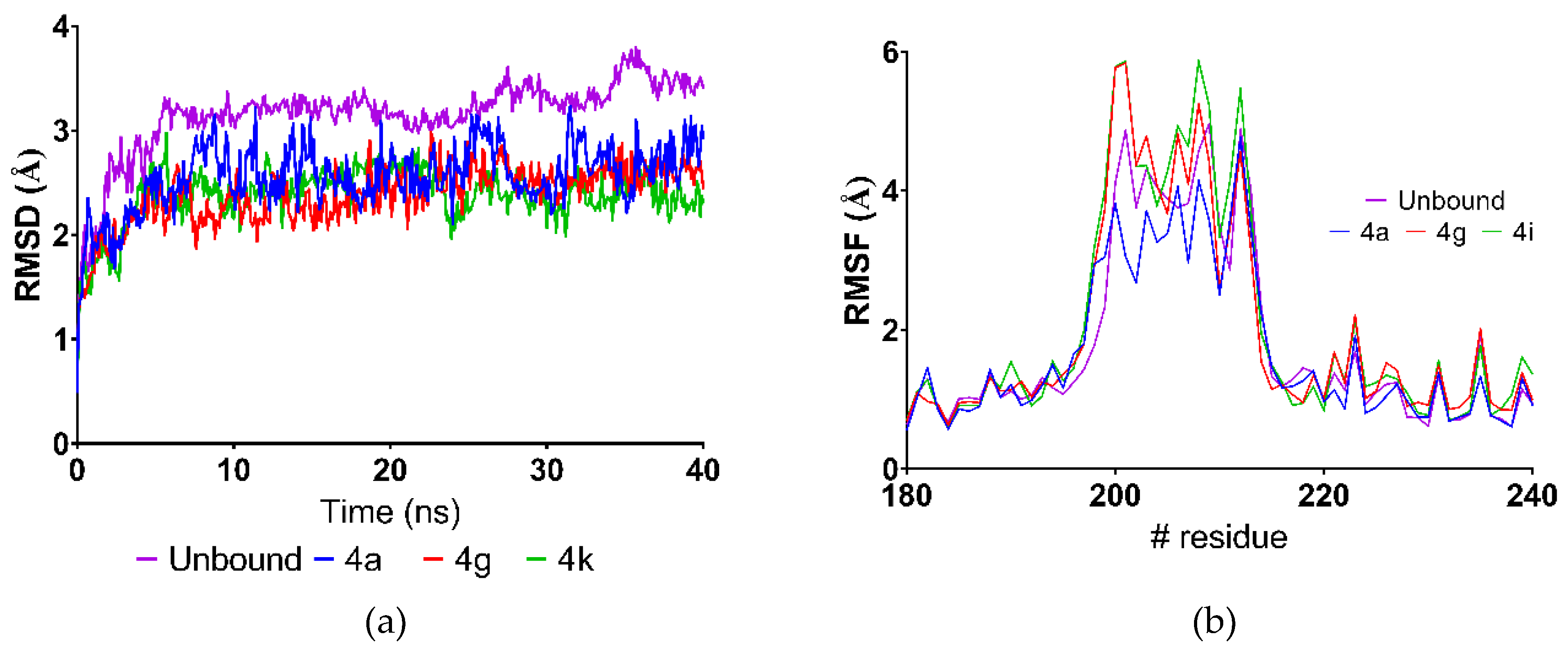
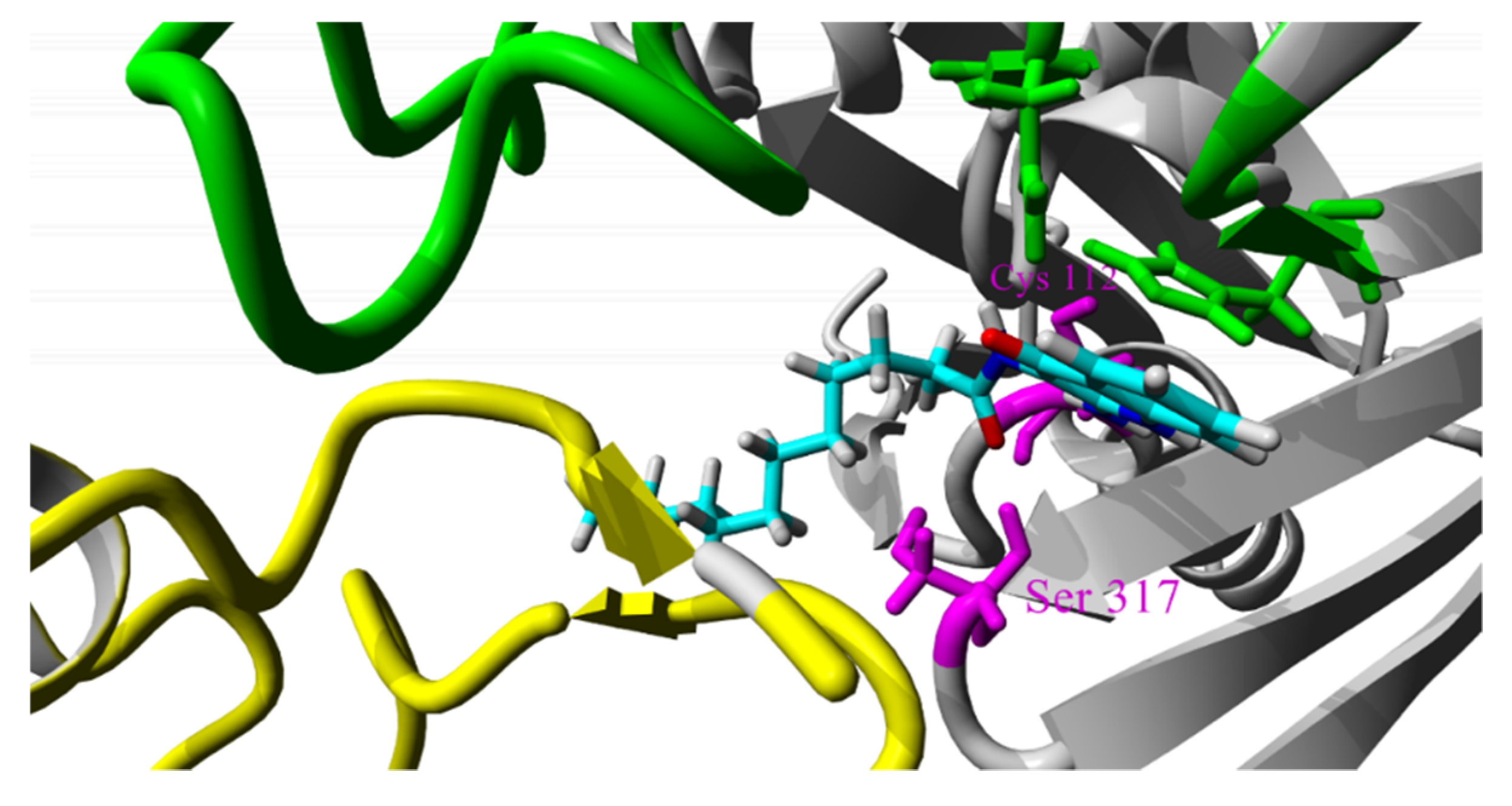
2.4. Molecular Dynamics Simulations
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. Evaluation of Biofilm Inhibition
4.3. Inhibition of Pyocyanin Formation
4.4. Molecular Docking
4.5. Molecular Dynamics Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Defoirdt, T.; Brackman, G.; Coenye, T. Quorum sensing inhibitors: How strong is the evidence? Trends Microbiol. 2013, 21, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Majik, M.S.; Parvatkar, P.T. Next generation biofilm inhibitors for Pseudomonas aeruginosa: Synthesis and rational design approaches. Curr. Top. Med. Chem. 2014, 14, 81–109. [Google Scholar] [CrossRef]
- Wilson, M. Bacterial biofilms and human disease. Sci. Prog. 2001, 84, 235–254. [Google Scholar] [CrossRef]
- Sharma, G.; Rao, S.; Bansal, A.; Dang, S.; Gupta, S.; Gabrani, R. Pseudomonas aeruginosa biofilm: Potential therapeutic targets. Biologicals 2014, 42, 1–7. [Google Scholar] [CrossRef]
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Kaufmann, G.F. Quo vadis quorum quenching? Curr. Opin. Pharmacol. 2013, 13, 688–698. [Google Scholar] [CrossRef]
- Blackwell, H.E.; Fuqua, C. Introduction to Bacterial Signals and Chemical Communication. Chem. Rev. 2011, 111, 1–3. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Storz, M.P.; Maurer, C.K.; Zimmer, C.; Wagner, N.; Brengel, C.; de Jong, J.C.; Lucas, S.; Müsken, M.; Häussler, S.; Steinbach, A.; et al. Validation of PqsD as an Anti-biofilm Target in Pseudomonas aeruginosa by Development of Small-Molecule Inhibitors. J. Am. Chem. Soc. 2012, 134, 16143–16146. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.T.; Noto, J.G.; Nichols-O’Neill, L.; Perez, L.J. Potent Irreversible Inhibitors of LasR Quorum Sensing in Pseudomonas aeruginosa. ACS Med. Chem. Lett. 2015, 6, 162–167. [Google Scholar] [CrossRef]
- Kalia, M.; Singh, P.K.; Yadav, V.K.; Yadav, B.S.; Sharma, D.; Narvi, S.S.; Mani, A.; Agarwal, V. Structure based virtual screening for identification of potential quorum sensing inhibitors against LasR master regulator in Pseudomonas aeruginosa. Microb. Pathog. 2017, 107, 136–143. [Google Scholar] [CrossRef]
- Sahner, J.H.; Empting, M.; Kamal, A.; Weidel, E.; Groh, M.; Börger, C.; Hartmann, R.W. Exploring the chemical space of ureidothiophene-2-carboxylic acids as inhibitors of the quorum sensing enzyme PqsD from Pseudomonas aeruginosa. Eur. J. Med. Chem. 2015, 96, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Sahner, J.H.; Brengel, C.; Storz, M.P.; Groh, M.; Plaza, A.; Müller, R.; Hartmann, R.W. Combining in Silico and Biophysical Methods for the Development of Pseudomonas aeruginosa Quorum Sensing Inhibitors: An Alternative Approach for Structure-Based Drug Design. J. Med. Chem. 2013, 56, 8656–8664. [Google Scholar] [CrossRef]
- Zhou, Z.; Ma, S. Recent Advances in the Discovery of PqsD Inhibitors as Antimicrobial Agents. ChemMedChem 2017, 12, 420–425. [Google Scholar] [CrossRef]
- Schütz, C.; Empting, M. Targeting the Pseudomonas quinolone signal quorum sensing system for the discovery of novel anti-infective pathoblockers. Beilstein J. Org. Chem. 2018, 14, 2627–2645. [Google Scholar] [CrossRef] [PubMed]
- Basak, A.; Abouelhassan, Y.; Kim, Y.S.; Norwood, V.M.; Jin, S.; Huigens, R.W. Halogenated quinolines bearing polar functionality at the 2-position: Identification of new antibacterial agents with enhanced activity against Staphylococcus epidermidis. Eur. J. Med. Chem. 2018, 155, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Zaheer, Z.; Khan, F.A.K.; Sangshetti, J.N.; Patil, R.H.; Lohar, K.S. Novel amalgamation of phthalazine–quinolines as biofilm inhibitors: One-pot synthesis, biological evaluation and in silico ADME prediction with favorable metabolic fate. Bioorg. Med. Chem. Lett. 2016, 26, 1696–1703. [Google Scholar] [CrossRef] [PubMed]
- Zuo, R.; Garrison, A.T.; Basak, A.; Zhang, P.; Huigens, R.W.; Ding, Y. In vitro antifungal and antibiofilm activities of halogenated quinoline analogues against Candida albicans and Cryptococcus neoformans. Int. J. Antimicrob. Agents 2016, 48, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Abouelhassan, Y.; Garrison, A.T.; Burch, G.M.; Wong, W.; Norwood, V.M.; Huigens, R.W. Discovery of quinoline small molecules with potent dispersal activity against methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis biofilms using a scaffold hopping strategy. Bioorg. Med. Chem. Lett. 2014, 24, 5076–5080. [Google Scholar] [CrossRef] [PubMed]
- Speck-Planche, A.; Kleandrova, V.V.; Scotti, M.T. Fragment-based approach for the in silico discovery of multi-target insecticides. Chemom. Intell. Lab. Syst. 2012, 111, 39–45. [Google Scholar] [CrossRef]
- Aleksić, I.; Šegan, S.; Andrić, F.; Zlatović, M.; Moric, I.; Opsenica, D.M.; Senerovic, L. Long-Chain 4-Aminoquinolines as Quorum Sensing Inhibitors in Serratia marcescens and Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 1425–1434. [Google Scholar] [CrossRef]
- Maurino-Reyes, E.D.J.; Gonzalez-Rodriguez, E.; Reyes-Rangel, F.; Lira-Rocha, A.; Loza-Mejia, M.A. A direct synthetic route to fused tricyclic quinolones from 2,3-diaminoquinolin-4(1H)one. Heterocycl. Commun. 2016, 22, 169–173. [Google Scholar] [CrossRef]
- Peeters, E.; Hooyberghs, G.; Robijns, S.; Waldrant, K.; De Weerdt, A.; Delattin, N.; Liebens, V.; Kucharíková, S.; Tournu, H.; Verstraeten, N.; et al. Modulation of the Substitution Pattern of 5-Aryl-2-Aminoimidazoles Allows Fine-Tuning of Their Antibiofilm Activity Spectrum and Toxicity. Antimicrob. Agents Chemother. 2016, 60, 6483–6497. [Google Scholar] [CrossRef] [Green Version]
- Abad-Zapatero, C.; Metz, J.T. Ligand efficiency indices as guideposts for drug discovery. Drug Discov. Today 2005, 10, 464–469. [Google Scholar] [CrossRef]
- Advanced Chemistry Development ChemSketch. Available online: www.acdlabs.com (accessed on 1 December 2018).
- Verdonk, M.L.; Rees, D.C. Group Efficiency: A Guideline for Hits-to-Leads Chemistry. ChemMedChem 2008, 3, 1179–1180. [Google Scholar] [CrossRef] [PubMed]
- Cavalluzzi, M.M.; Mangiatordi, G.F.; Nicolotti, O.; Lentini, G. Ligand efficiency metrics in drug discovery: The pros and cons from a practical perspective. Expert Opin. Drug Discov. 2017, 12, 1087–1104. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.-S.; Ok, K.; Kim, Y.; Park, H.-D.; Byun, Y. Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 2913–2917. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, J.T.; Galloway, W.R.J.D.; Wright, M.; Mati, I.K.; Nicholson, R.L.; Welch, M.; Spring, D.R. Design, synthesis and biological evaluation of non-natural modulators of quorum sensing in Pseudomonas aeruginosa. Org. Biomol. Chem. 2012, 10, 6032–6044. [Google Scholar] [CrossRef] [PubMed]
- Leeson, P.D.; Springthorpe, B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 2007, 6, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Muraglia, E.; Bazzo, R.; Carfì, A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, M.C.; Dong, S.-H.; Rossi, F.M.; Karlen, K.M.; Kumar, R.S.; Nair, S.K.; Blackwell, H.E. Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop “Out” Conformation for LasR. Cell Chem. Biol. 2018, 25, 1128–1139.e3. [Google Scholar] [CrossRef]
- Bera, A.K.; Atanasova, V.; Robinson, H.; Eisenstein, E.; Coleman, J.P.; Pesci, E.C.; Parsons, J.F. Structure of PqsD, a Pseudomonas Quinolone Signal Biosynthetic Enzyme, in Complex with Anthranilate. Biochemistry 2009, 48, 8644–8655. [Google Scholar] [CrossRef]
- Steinbach, A.; Maurer, C.K.; Weidel, E.; Henn, C.; Brengel, C.; Hartmann, R.W.; Negri, M. Molecular basis of HHQ biosynthesis: Molecular dynamics simulations, enzyme kinetic and surface plasmon resonance studies. BMC Biophys. 2013, 6, 10. [Google Scholar] [CrossRef]
- Cruciata, M.; Gaglio, R.; Scatassa, M.L.; Sala, G.; Cardamone, C.; Palmeri, M.; Moschetti, G.; La Mantia, T.; Settanni, L. Formation and Characterization of Early Bacterial Biofilms on Different Wood Typologies Applied in Dairy Production. Appl. Environ. Microbiol. 2018, 84, e02107-17. [Google Scholar] [CrossRef]
- Pires, D.; Sillankorva, S.; Faustino, A.; Azeredo, J. Use of newly isolated phages for control of Pseudomonas aeruginosa PAO1 and ATCC 10145 biofilms. Res. Microbiol. 2011, 162, 798–806. [Google Scholar] [CrossRef] [Green Version]
- Kwasny, S.M.; Opperman, T.J. Static biofilm cultures of Gram-positive pathogens grown in a microtiter format used for anti-biofilm drug discovery. Curr. Protoc. Pharmacol. 2010, 50, 13A.8.1–13A.8.23. [Google Scholar] [CrossRef]
- Martínez Díaz, Y.; Vanegas Laverde, G.; Reina Gamba, L.; Mayorga Wandurraga, H.; Arévalo-Ferro, C.; Ramos Rodríguez, F.; Duque Beltrán, C.; Castellanos Hernández, L. Biofilm inhibition activity of compounds isolated from two Eunicea species collected at the Caribbean Sea. Rev. Bras. Farmacogn. 2015, 25, 605–611. [Google Scholar] [CrossRef] [Green Version]
- Tapia-Rodriguez, M.R.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez-Tellez, M.A.; Martins, C.M.; Ayala-Zavala, J.F. Carvacrol as potential quorum sensing inhibitor of Pseudomonas aeruginosa and biofilm production on stainless steel surfaces. Food Control. 2017, 75, 255–261. [Google Scholar] [CrossRef]
- Wavefunction Inc Spartan ’10. Available online: www.wavefun.com (accessed on 1 December 2018).
- Thomsen, R.; Christensen, M.H. MolDock: A New Technique for High-Accuracy Molecular Docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- CLC Bio Molegro Virtual Docker. Available online: https://www.qiagenbioinformatics.com/ (accessed on 1 December 2018).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasara Dynamics. Available online: www.yasara.org (accessed on 1 December 2018).
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 3a-i and 4a-i are available from the authors. |
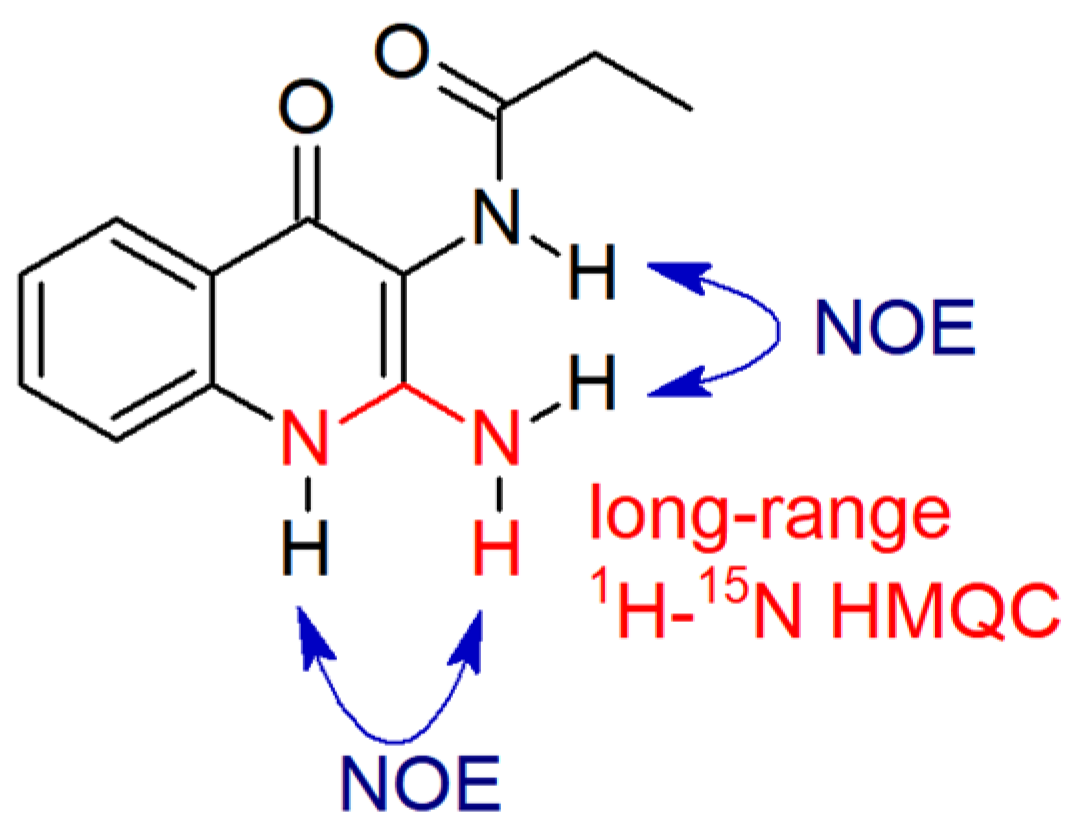

| BI (%) (±s.d.) | BI (%) (±s.d.) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cmp | n | S.a | P.a | PEI | Cmp | n | S.a | P.a | PEI |
| 3a | 1 | 15.3 (±2.8) | 26.4 (±4.6) | 1.22 | 4a | 1 | < 5 | 28.65 (±2.2) | 1.24 |
| 3b | 2 | 31.2 (±3.6) | 30.6 (±3.9) | 1.33 | 4b | 2 | 29.8 (±5.1) | 29.43 (±3.1) | 1.20 |
| 3c | 3 | <5 | 37.5 (±2.6) | 1.54 | 4c | 3 | <5 | 33.09 (±3.2) | 1.28 |
| 3d | 4 | <5 | 36.6 (±1.7) | 1.42 | 4d | 4 | <5 | 44.95 (±1.6) | 1.64 |
| 3e | 6 | 22.6 (±5.7) | 43.8 (±3.4) | 1.53 | 4e | 6 | <5 | 48.97 (±4.6) | 1.62 |
| 3f | 8 | 24.8 (±1.5) | 50.6 (±4.0) | 1.61 | 4f | 8 | 11.9 (±4.2) | 53.38 (±2.5) | 1.62 |
| 3g | 10 | 18.0 (±1.2) | 54.2 (±4.0) | 1.46 | 4g | 10 | 15.7 (±5.8) | 61.45 (±2.2) | 1.72 |
| 3h | 12 | 25.7 (±1.1) | 52.3 (±4.2) | 1.41 | 4h | 12 | 16.8 (±4.4) | 57.63 (±3.2) | 1.49 |
| 3i | 14 | 34.7 (±2.0) | 49.8 (±1.8) | 1.25 | 4i | 14 | 14.2 (±5.4) | 56.62 (±3.1) | 1.37 |
| MolDock Score | MolDock Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligand | BI (%) | PqsD | LasR | logP | Ligand | BI (%) | PqsD | LasR | logP |
| 3a | 26.36 | −99.478 | −101.21 | 2.53 | 4a | 28.65 | −96.4705 | −104.98 | 1.11 |
| 3b | 30.61 | −103.646 | −109.25 | 3.06 | 4b | 29.43 | −104.784 | −110.48 | 1.64 |
| 3c | 37.54 | −105.508 | −113.46 | 3.59 | 4c | 33.09 | −110.356 | −116.89 | 2.17 |
| 3d | 36.56 | −111.982 | −114.62 | 4.12 | 4d | 44.95 | −117.802 | −125.21 | 2.70 |
| 3e | 43.84 | −125.746 | −137.16 | 5.18 | 4e | 48.97 | −129.125 | −142.73 | 3.76 |
| 3f | 50.57 | −131.982 | −152.30 | 6.24 | 4f | 53.38 | −136.142 | −145.82 | 4.82 |
| 3g | 54.20 | −124.547 | −151.66 | 6.77 | 4g | 61.45 | −139.494 | −152.81 | 5.35 |
| 3h | 52.27 | −125.681 | −146.32 | 7.30 | 4h | 57.63 | −133.275 | −159.79 | 5.88 |
| 3i | 49.82 | −147.112 | −156.07 | 7.83 | 4i | 56.62 | −150.072 | −181.92 | 6.41 |
| LasR | PqsD | |||||
|---|---|---|---|---|---|---|
| Average RMSD (Å) | LBE (KJ/mol) | GE 2 | Average RMSD (Å) | LBE (KJ/mol) | GE 2 | |
| 4a | 1.79 | 7.09 | -- | 2.55 | 75.69 | -- |
| 4g | 2.01 | 2.01 | −0.56 | 2.38 | 200.44 | 13.86 |
| 4i | 1.70 | −11.26 | −1.41 | 2.37 | 229.30 | 11.81 |
| AHL | 1.76 | 3.29 | -- | 3.26 | ---- | -- |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinosa-Valdés, M.P.; Borbolla-Alvarez, S.; Delgado-Espinosa, A.E.; Sánchez-Tejeda, J.F.; Cerón-Nava, A.; Quintana-Romero, O.J.; Ariza-Castolo, A.; García-Del Río, D.F.; Loza-Mejía, M.A. Synthesis, In Silico, and In Vitro Evaluation of Long Chain Alkyl Amides from 2-Amino-4-Quinolone Derivatives as Biofilm Inhibitors. Molecules 2019, 24, 327. https://doi.org/10.3390/molecules24020327
Espinosa-Valdés MP, Borbolla-Alvarez S, Delgado-Espinosa AE, Sánchez-Tejeda JF, Cerón-Nava A, Quintana-Romero OJ, Ariza-Castolo A, García-Del Río DF, Loza-Mejía MA. Synthesis, In Silico, and In Vitro Evaluation of Long Chain Alkyl Amides from 2-Amino-4-Quinolone Derivatives as Biofilm Inhibitors. Molecules. 2019; 24(2):327. https://doi.org/10.3390/molecules24020327
Chicago/Turabian StyleEspinosa-Valdés, Mariana Paola, Sara Borbolla-Alvarez, Ana Elena Delgado-Espinosa, Juan Francisco Sánchez-Tejeda, Anabelle Cerón-Nava, Osvaldo Javier Quintana-Romero, Armando Ariza-Castolo, Diego Fernando García-Del Río, and Marco A. Loza-Mejía. 2019. "Synthesis, In Silico, and In Vitro Evaluation of Long Chain Alkyl Amides from 2-Amino-4-Quinolone Derivatives as Biofilm Inhibitors" Molecules 24, no. 2: 327. https://doi.org/10.3390/molecules24020327