Novel Thiazolidin-4-ones as Potential Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Rational Design of the Compounds
2.2. Computational Prediction of Anti-HIV Activity
2.3. Molecular Docking Prediction
2.4. Prediction of Toxicity
2.5. Chemistry
2.6. HIV-1 RT Inhibitory Action
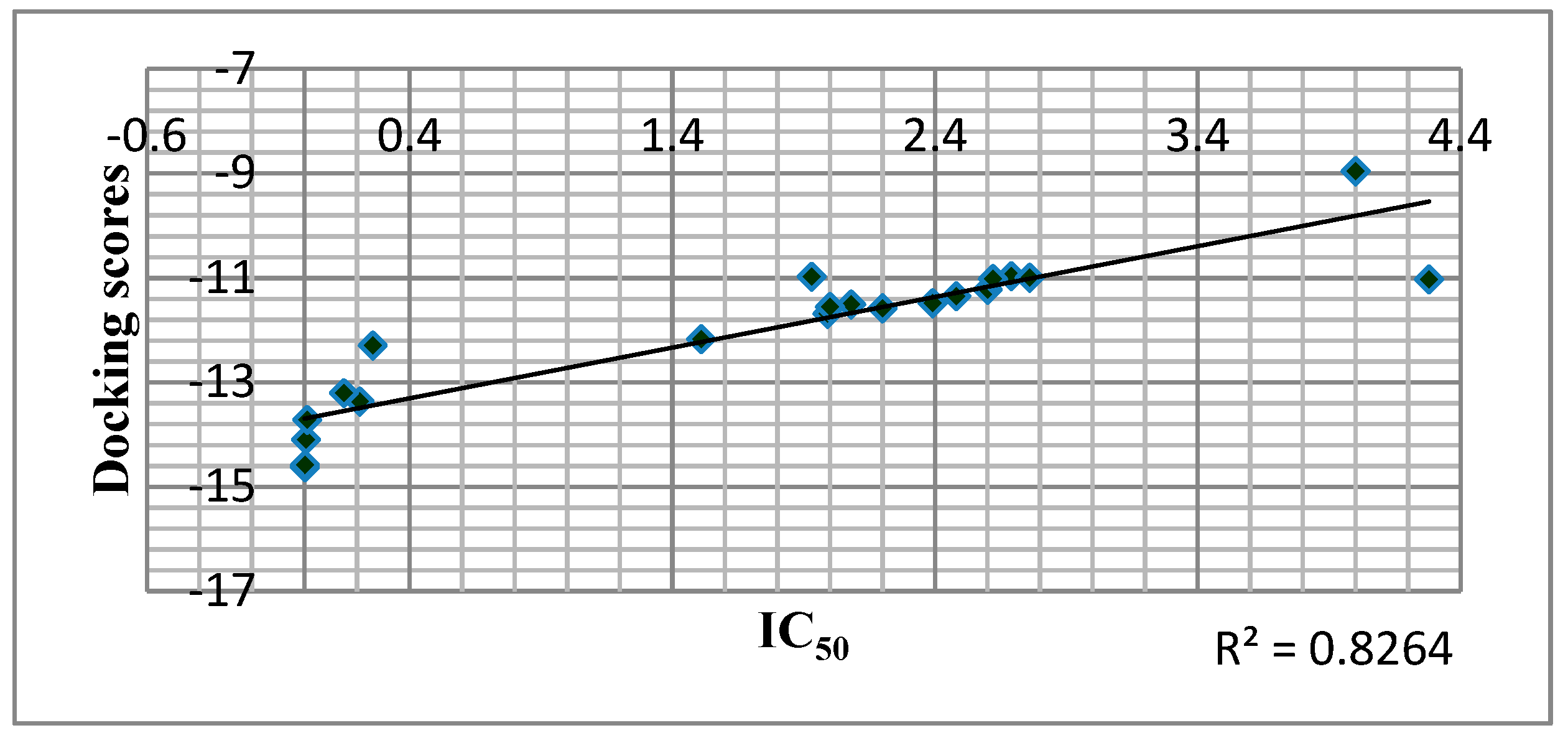
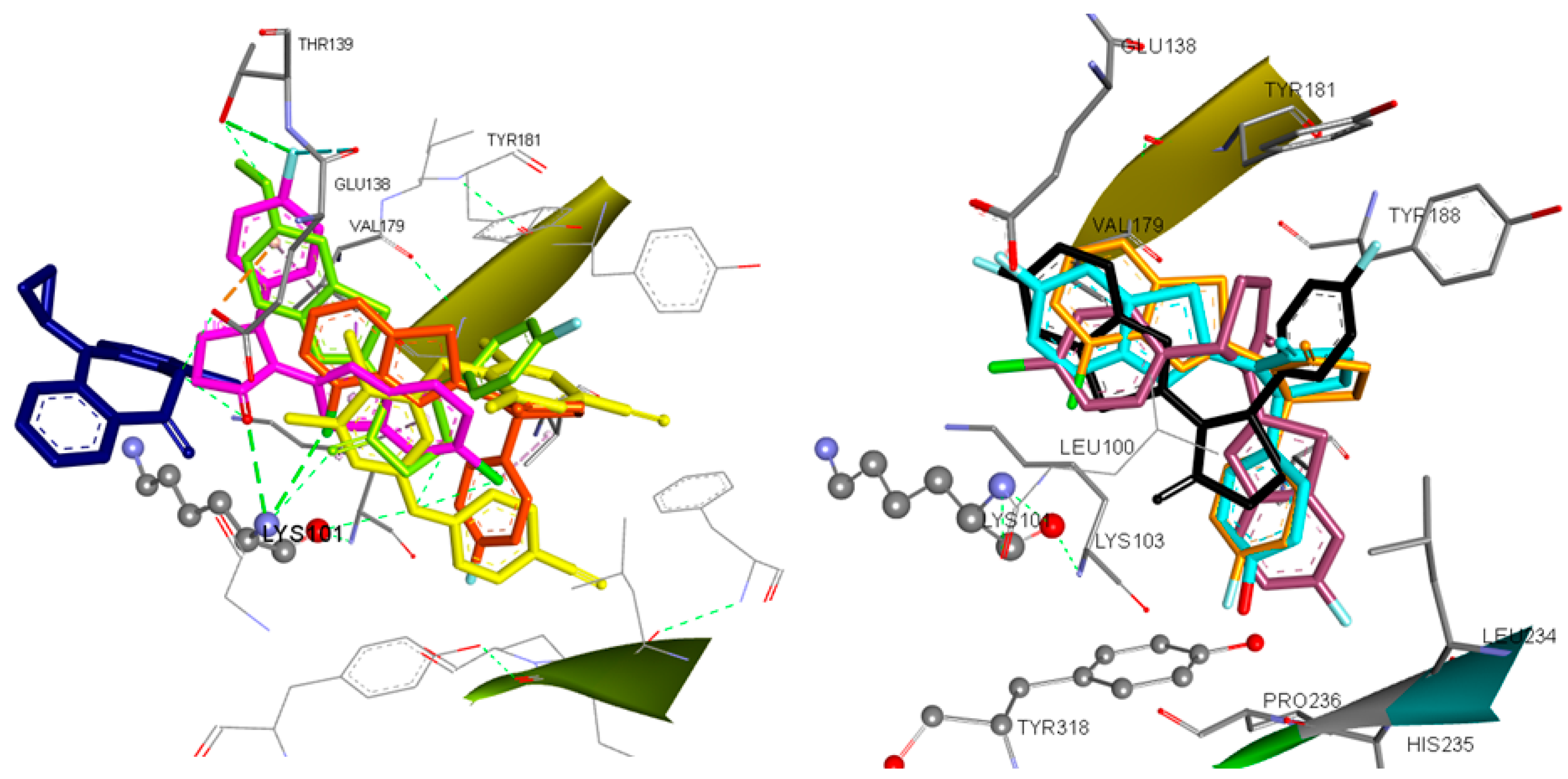
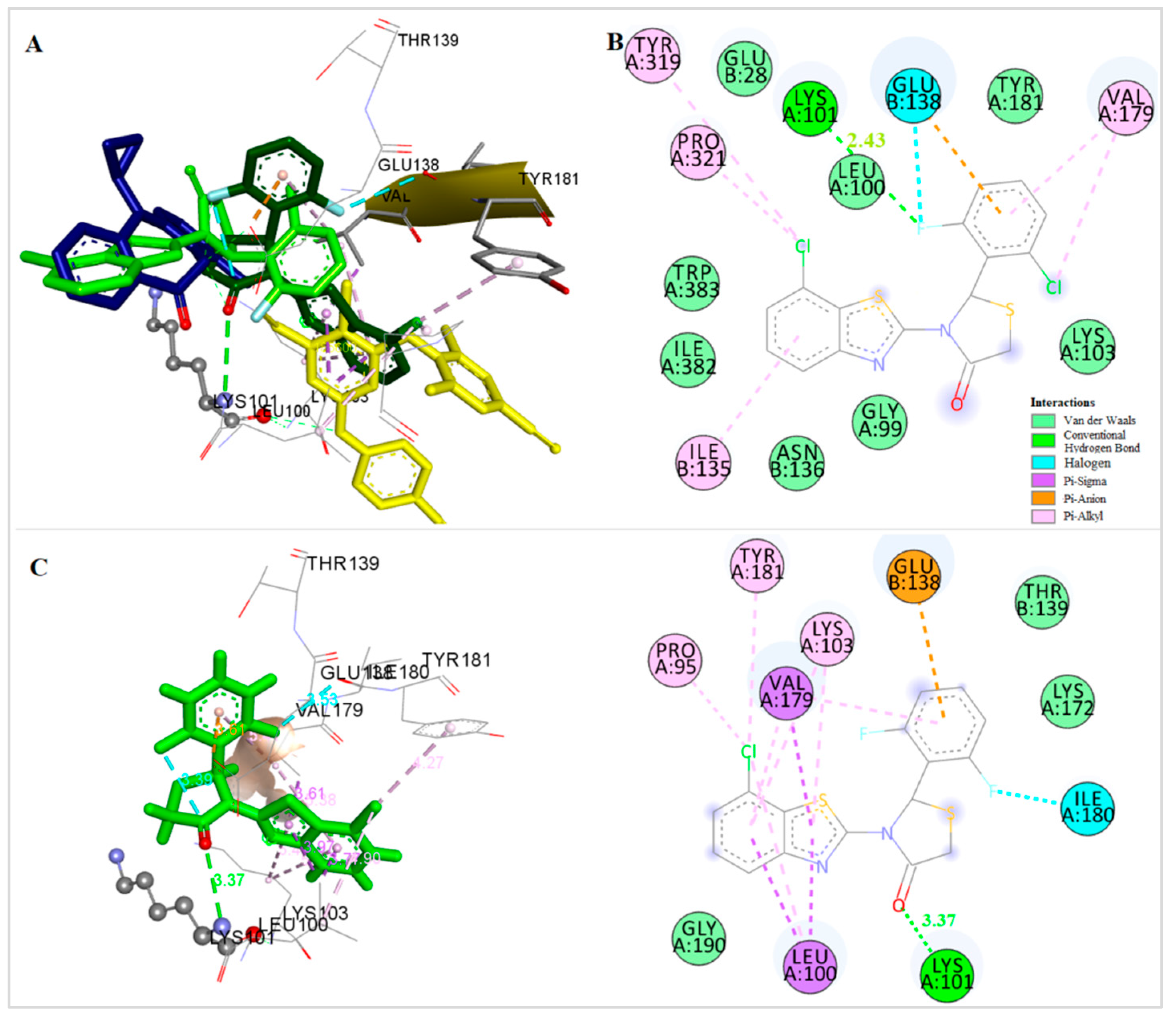
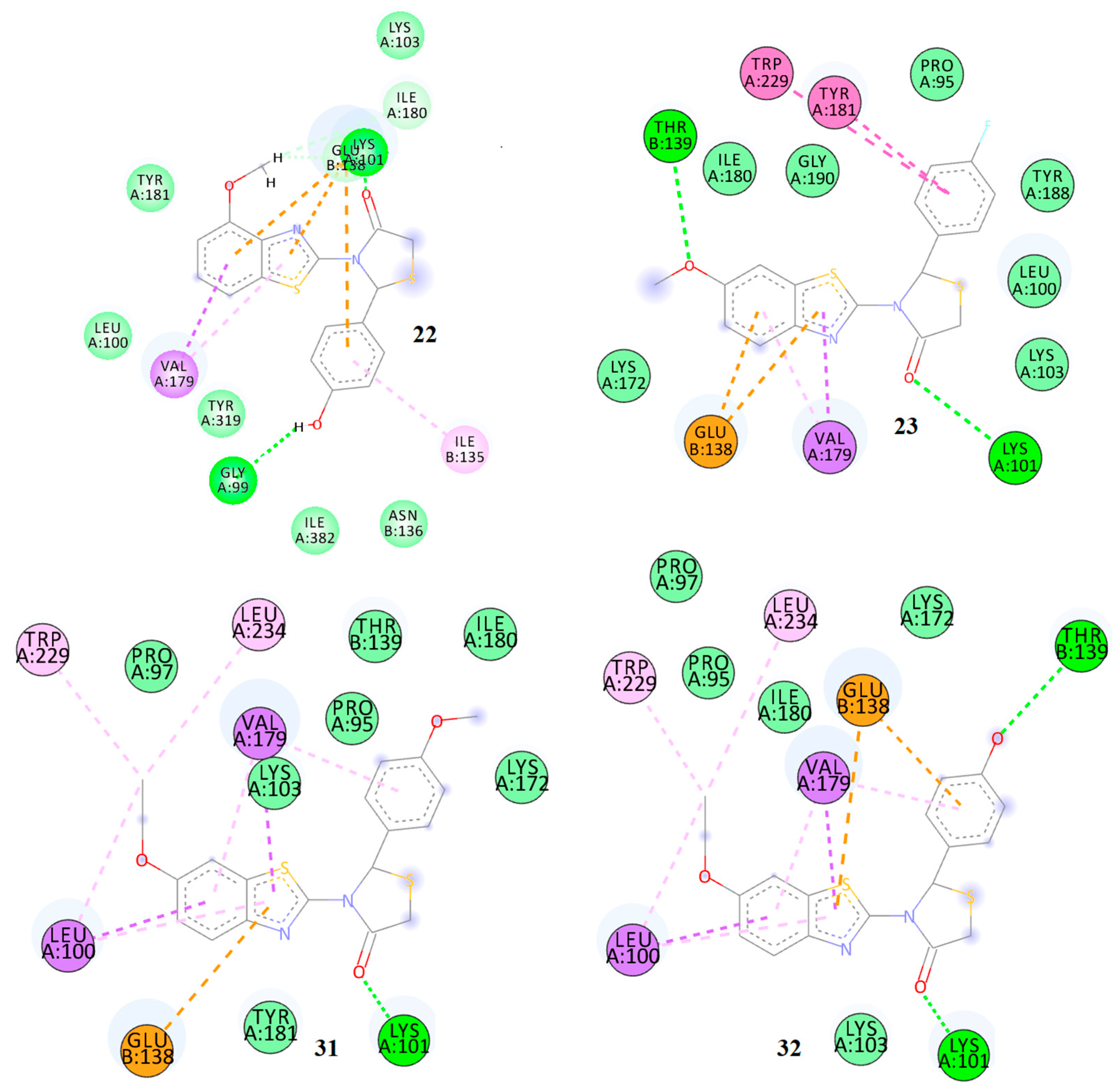
2.7. Docking Analysis
2.8. Cytotoxicity Assessment
3. Materials and Methods
3.1. Computer Simulation Methods
3.2. PASS Prediction
3.3. Prediction of Toxicity
3.4. Chemistry
3.4.1. General Procedure for the Synthesis of Thiazolidinones by Conventional Method
3.4.2. Microwave Irradiation Experiments
3.4.3. Synthesis of 3-(7-Chloro-benzo[d]thiazol-2-yl)-2-(2,6-difluorophenyl)thiazolidin-4-one (1)
3.4.4. Synthesis of 3-(7-Chloro-benzo[d]thiazol-2-yl)-2-(2-chloro-6-fluorophenyl)thiazolidin-4-one (2)
3.4.5. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (3)
3.4.6. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (4)
3.4.7. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (5)
3.4.8. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (6)
3.4.9. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (7)
3.4.10. Synthesis of 3-(6-Chloro-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (8)
3.4.11. Synthesis of 3-(6-Chloro-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (9)
3.4.12. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (10)
3.4.13. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (11)
3.4.14. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (12)
3.4.15. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (13)
3.4.16. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (14)
3.4.17. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-flurophenyl)thiazolidin-4-one (15)
3.4.18. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (16)
3.4.19. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (17)
3.4.20. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (18)
3.4.21. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (19)
3.4.22. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (20)
3.4.23. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (21)
3.4.24. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (22)
3.4.25. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (23)
3.4.26. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (24)
3.4.27. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (25)
3.4.28. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (26)
3.4.29. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (27)
3.4.30. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (28)
3.4.31. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(2,6-dichlorophenyl) thiazolidin-4-one (29)
3.4.32. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(2,3-dichlorophenyl) thiazolidin-4-one (30)
3.4.33. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (31)
3.4.34. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4- one (32)
3.5. Evaluation of RT Inhibitory Action
3.6. Toxicity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ravichandran, S.; Veerasamy, F.; Raman, S.; Krishnan, P.N.; Agrawal, R.K. An overview on HIV-1 reverse transcriptase inhibitors. Dig. J. Nanomater. Biostructures 2008, 3, 171–187. [Google Scholar]
- Asahchop, E.L.; Wainberg, M.A.; Sloan, R.D.; Tremblay, C.L. Antiviral drug resistance and the need for development of new HIV-1 reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 2012, 56, 5000–5008. [Google Scholar] [CrossRef] [PubMed]
- Reeves, J.D.; Doms, R.W. Human immunodeficiency virus type 2. J. Gen. Virol. 2002, 83, 1253–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andries, K.; Azijn, H.; Thielemans, T.; Ludovici, D.; Kukla, M.; Heeres, J.; Janssen, P.; De Corte, B.; Vingerhoets, J.; Pauwels, R.; et al. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2004, 48, 4680–4686. [Google Scholar] [CrossRef]
- Kaushik, N.K.; Kaushik, N.; Attri, P.; Kumar, N.; Kim, C.H.; Verma, A.K.; Choi, E.H. Biomedical importance of indoles. Molecules 2013, 18, 6620–6662. [Google Scholar] [CrossRef]
- Kadow, J.F.; Regueiro-Ren, A.; Xue, Q.M. Indole, Azaindole and Related Heterocyclic Sulfonylureido Piperazine Derivatives. US Patent 6,900,206, 9 June 2003. [Google Scholar]
- Pauwels, R.; Andries, K.; Debyser, Z.; Van Daele, P.; Schols, D.; Stoffels, P.; De Vreese, K.; Woestenborghs, R.; Vandamme, A.M.; Janssen, C.G. Potent and highly selective human immunodeficiency virus type 1 (HIV-1) inhibition by a series of alpha-anilinophenylacetamide derivatives targeted at HIV-1 reverse transcriptase. Proc. Nat. Acad. Sci. USA 1993, 90, 1711–1717. [Google Scholar] [CrossRef]
- Pauwels, R.; Andries, K.; Desmyter, J.; Schols, D.; Kukla, M.J.; Breslin, H.J.; Raeymaeckers, A.; VanGelder, J.; Woestenborghs, R.; Heykants, J.; et al. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBOderivatives. Nature 1990, 343, 470–474. [Google Scholar] [CrossRef]
- Debyser, Z.; Pauwels, R.; Andries, K.; Desmyter, J.; Kukla, M.; Janssen, P.A.; De Clercq, E. An antiviral target on reverse transcriptase of human immunodeficiency virus type 1 revealed bytetrahydroimidazo-[4,5,1-jk][1,4]benzodiazepin-2 (1H)-one and -thione derivatives. Proc. Nat. Acad. Sci. USA 1991, 88, 1451–1455. [Google Scholar] [CrossRef]
- Le Van, K.; Cauvin, C.; Walque, S.; Georges, B.; Boland, S.; Martinelli, V.; Demonté, D.; Durant, F.; Hevesi, L.; Van Lint, C. New pyridinone derivatives as potent HIV-1 nonnucleoside reverse transcriptase inhibitors. J. Med. Chem 2009, 52, 3636–3643. [Google Scholar] [CrossRef]
- Tanaka, H.; Takashima, H.; Ubasawa, M.; Sekiya, K.; Inouye, N.; Baba, M.; Shigeta, S.; Walker, R.T.; De Clercq, E.; Miyasaka, T.J. Synthesis and antiviral activity of 6-benzyl analogs of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT)as potent and selective anti-HIV-1 agents. Med. Chem. 1995, 38, 2860–2865. [Google Scholar] [CrossRef]
- Penta, A.; Ganguly, S.; Murugesan, S. Design and synthesis of tetrahydrophthalimide derivatives as inhibitors of HIV-1 reverse transcriptase. Org. Med. Chem. Lett. 2013, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Rawal, R.K.; Tripathi, R.; Kulkarni, S.; Paranjape, R.; Katti, S.B.; Pannecouque, C.; De Clercq, E. 2-(2,6-Dihalo-phenyl)-3-heteroaryl-2-ylmethyl-1, 3-thiazolidin-4-ones: Anti-HIVagents. Chem. Biol. Drug Des. 2008, 72, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Khalid, Z.; Aslam, S.; Ahmad, M.; Munawar, A.; Montero, K.; Detorio, M.; Parvez, M.; Schinazi, R.F. Anti-HIV activity of new pyrazolobenzothiazine 5,5-dioxide-based acetohydrazides. Med. Chem. Res. 2015, 24, 3671–3680. [Google Scholar] [CrossRef]
- Novikov, M.S.; Valuev-Elliston, V.T.; Babkov, D.A.; Paramonova, M.P.; Ivanov, A.V.; Gavryushov, S.A.; Khandazhinskaya, A.L.; Kochetkov, S.N.; Pannecouque, C.; Andrei, G.; et al. N1,N3-disubstituted uracils as nonnucleosides inhibitors of HIV-1 reverse transcriptase. Bioorg. Med. Chem. 2013, 21, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Chen, F.-E.; De Clercq, E.; Balzarini, J.; Pannecouque, C. Synthesis and in vitroanti-HIV evaluation of a new series of 6-arylmethyl-substituted S-DABOs as potentialnon-nucleoside HIV-1 reverse transcriptase inhibitors. Eur. J. Med. Chem. 2009, 41, 1016–1023. [Google Scholar] [CrossRef]
- Pitta, E.; Tsolaki, E.; Geronikaki, A.; Petrović, J.; Glamočlija, J.; Soković, M.; Crespan, E.; Maga, G.; Bhunia, S.S.; Saxena, A.K. 4-Thiazolidinone derivatives as potent antimicrobial agents: Microwave-assisted synthesis, biological evaluation and docking studies. MedChemComm. 2015, 6, 319–326. [Google Scholar] [CrossRef]
- Zahid, M.; Yasin, K.A.; Akhtar, T.; Rama, N.H.; Hameed, S.; Al-Masoudi, N.A.; Loddo, R.; Colla, P. Synthesis and in vitro antiproliferative activity of new adamantylthiazolyl-1,3,4-oxadiazoles. ARKIVOC 2009, xi, 85–93. [Google Scholar]
- La Regina, G.; Coluccia, A.; Piscitelli, F.; Bergamini, A.; Sinistro, A.; Cavazza, A.; Maga, J.; Samuele, A.; Zanoli, S.; Novellino, E.; et al. Indolyl aryl sulfones as HIV-1 non-nucleoside reverse transcriptase inhibitors: Role of two halogen atoms at the indole ring in developing new analogues with improved antiviral activity. J. Med. Chem. 2007, 50, 5034–5038. [Google Scholar] [CrossRef]
- Akkouh, O.; Ng, T.B.; Singh, S.S.; Yin, C.; Dan, X.; Chan, Y.S.; Pan, W.; Cheung, R.C.F. Lectins with Anti-HIV activity: A review. Molecules 2015, 20, 648–668. [Google Scholar] [CrossRef]
- Famiglini, V.; Coluccia, A.; Brancale, A.; Pelliccia, S.; La Regina, G.; Silvestri, R. Arylsulfone-based HIV-I non-nucleoside reverse transcriptase inhibitors. Future. Med. Chem. 2013, 5, 2141–2156. [Google Scholar] [CrossRef]
- De Clercq, E. Dancing with chemical formulae of antivirals: A personal account. Biochem. Pharm. 2013, 86, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Veljkovic, N.; Glisic, S.; Prljic, J.; Perovic, V.; Veljkovic, V. Simple and general criterionfor “in silico” screening of candidate HIV drugs. Curr. Pharm. Biotechnol. 2013, 14, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhan, P.; Liu, H.; Pannecouque, C.; Balzarini, J.; De Clercq, E. Synthesis and biological evaluation of pyrazine derivatives as novel HIV-1 NNRTIs. Bioorg. Med. Chem. 2013, 21, 2128–2134. [Google Scholar] [CrossRef] [PubMed]
- La Regina, G.; Coluccia, A.; Brancale, A.; Piscitelli, F.; Gatti, V.; Maga, G.; Sa-muele, A.; Pannecouque, C.; Schols, D.; Balzarini, J.; et al. Indolylarylsulfones as HIV-1 non-nucleoside reverse transcriptase inhibitors: New cyclic substituents at indole-2-carboxamide. J. Med. Chem. 2011, 54, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- La Regina, G.; Coluccia, A.; Brancale, A.; Piscitelli, F.; Famiglini, V.; Cosconati, S.; Maga, G.; Samuele, A.; Gonzales, E.; Clotet, B.; et al. New nitrogen containing substituents at the indole-2-carboxamide yield high potent and broad spectrum indolylarylsulfone HIV-1 non-nucleoside reverse transcriptase inhibitors. J. Med. Chem. 2012, 55, 6634–6638. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Cheng, X.; Sun, L.; Song, S.; Álvarez, M.; Luczkowiak, J.; Pannecouque, C.; De Clercq, E.; Menéndez-Arias, L.; Zhan, P.; et al. Design, synthesis and biological evaluation of 3-hydroxyquinazoline-2,4(1H,3H)-diones as dual inhibitors of HIV-1 reverse transcriptase-associated RNase H and integrase. Bioorg. Med. Chem. 2019, 27, 3836–3845. [Google Scholar] [CrossRef]
- Tian, Y.; Zhan, P.; Rai, D.; Zhang, J.; Clercq, E.D.; Liu, X. Recent advances in the research of 2,3-diaryl-1,3-thiazolidin-4-one derivatives as potent HIV-1 non-nucleoside reverse transcriptase inhibitors. Curr. Med. Chem. 2012, 19, 2026–2037. [Google Scholar] [CrossRef]
- Suryawanshi, R.; Jadhav, S.; Makwana, N.; Desai, D.; Chaturbhuj, D.; Sonawani, A.; Idicula-Thomas, S.; Murugesan, V.; Katti, S.; Tripathy, S.; et al. Evaluation of 4-thiazolidinone derivatives as potential reverse transcriptase inhibitors against HIV-1 drug resistant strains. Bioorg. Chem. 2017, 71, 211–218. [Google Scholar] [CrossRef]
- Rao, A.; Balzarini, J.; Carbone, A.; Chimirri, A.; De Clercq, E.; Monforte, A.; Monforte, P.; Pannecouque, C.; Zappala, M. 2-(2,6-Dihalophenyl)-3-(pyrimidin-2-yl)-1,3-thiazolidin-4-ones as non-nucleoside HIV-1 reverse transcriptase inhibitors. Antivir. Res. 2004, 63, 79–84. [Google Scholar] [CrossRef]
- Chen, H.; Guo, Z.; Yin, Q.; Duan, X.; Gu, Y.; Li, X. Design, synthesis and HIV-RT inhibitory activity of novel thiazolidin-4-one derivatives. Front. Chem. Sci. Eng. 2011, 5, 231–237. [Google Scholar] [CrossRef]
- Rawal, K.; Prabhakar, Y.; Katti, B.V. Molecular surface features in modeling the HIV-1 RT inhibitory activity of 2-(2,6-disubstituted phenyl)-3-(substituted pyrimidin-2-yl)-thiazolidin-4-ones. QSAR Comb. Chem. Sci. 2007, 26, 398. [Google Scholar] [CrossRef]
- Kumar, S.; Purohit, D.; Pandey, P. Molecular docking and its application towards modern drug discovery. World J. Pharm. Pharm. Sci. 2019, 6, 691–696. [Google Scholar]
- Eweas, A.F.; Maghrabi, I.A.; Namarneh, A.I. Advances in molecular modeling and docking as a tool for modern drug discovery. Der Pharma Chem 2014, 6, 211–228. [Google Scholar]
- Zhou, Z.; Lin, X.; Madura, J.D. HIV-1 RT Nonnucleoside inhibitors and their interaction with RT for antiviral drug development. Infect. Disord. Drug Targets 2006, 6, 391–413. [Google Scholar] [CrossRef]
- De Clercq, E. Where rilpivirine meets with tenofovir, the start of a new anti-HIV drug combination era. Biochem. Pharm. 2012, 84, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskiy, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar]
- Geronikaki, A.; Dearden, J.C.; Filimonov, D.; Galaeva, I.; Garibova, T.L.; Gloriozova, T.; Kraineva, V.; Lagunin, A.; Macaev, F.Z.; Molodavkin, G.; et al. Design of new cognition enhancers: From computer prediction to synthesis and biological evaluation. J. Med. Chem. 2004, 47, 2870–2876. [Google Scholar] [CrossRef] [PubMed]
- Geronikaki, A.; Babaev, E.; Dearden, J.; Dehaen, W.; Filimonov, D.; Galaeva, I.; Kraineva, V.; Lagunin, A.; Macaev, F.; Molodavkin, G.; et al. Design, synthesis, computational and biological evaluation of new anxiolytics. Bioorg. Med. Chem. 2004, 12, 6559–6568. [Google Scholar] [CrossRef] [PubMed]
- Famiglini, V.; Silvestri, R. Focus on chilarity of HIV-1 non-nucleoside reverse transcriptase inhibitors. Molecules 2016, 21, 221. [Google Scholar] [CrossRef]
- Balzarini, J.; Orzeszko, B.; Maurin, J.; Orzerszko, A. Synthesis and anti-HIV studies of 2-adamantyl-substituted thiazolidin-4-ones. Eur. J. Med. Chem. 2007, 42, 993–1003. [Google Scholar] [CrossRef]
- Chimirri, A.; Grasso, S.; Molica, C.; Monforte, A.-M.; Monforte, P.; Zappalà, M.; De Clercq, E. Structural features and anti-human immunodeficiency virus (HIV) activity of the isomers of 1-(2′,6′-difluorophenyl)-1H,3H-thiazolo[3,4-a]benzimidazole, a potent non-nucleoside HIV-1 reverse transcriptase inhibitor. Antivir. Chem. Chemother. 1997, 8, 363–370. [Google Scholar] [CrossRef]
- Prajapati, D.; Ramajayam, R.; Ram Yadar, M.; Giridhar, R. The search for potent, small molecule NNRTIs: A review. Biorg. Med. Chem. 2009, 17, 5744–5762. [Google Scholar] [CrossRef] [PubMed]
- Pitta, E.; Geronikaki, A.; Surmava, S.; Eleftheriou, P.; Mehta, V.P.; Van der Eycken, E.V. Synthesis and HIV-1 RT inhibitory action of novel (4/6-substituted benzo[d]thiazol-2-yl)thiazolidin-4-ones. Divergence from the non-competitive inhibition mechanism. J. Enzyme Inhib. Med. Chem. 2013, 8, 113–122. [Google Scholar] [CrossRef]
- Kouatly, O.; Eleftheriou, P.; Petrou, A.; Hadjipavlou-Litina, D.; Geronikaki, A. Docking assisted design of novel 4-adamantanyl-2-thiazolylimino-5-arylidene-4-thiazolidinones as potent NSAIDs. SAR QSAR Environ. Res. 2018, 29, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.rcsb.org/structure/3MEC (accessed on 9 November 2016).
- Nadaraia, N.S.; Amiranashvili, L.S.; Merlani, M.; Kakhabrishvili, M.L.; Barbakadze, N.N.; Geronikaki, A.; Petrou, A.; Poroikov, V.; Ciric, A.; Glamoclija, J.; et al. Novel antimicrobial agents’ discovery among the steroid derivatives. Steroids 2019, 144, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Helma, C. Lazy structure-activity relationships (lazar) for the prediction of rodent carcinogenicity and Salmonella mutagenicity. Mol. Divers. 2006, 10, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Maunz, A.; Helma, C. Prediction of chemical toxicity with local support vector regression and activity-specific kernels. Sar. Qsar Environ. Res. 2008, 19, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Vizirianakis, I.S.; Tsiftsoglou, A.S. Blockade of murine erythroleukemia cell differentiation by hypomethylating agents causes accumulation of discrete small poly(A)- RNAs hybridized to 3′-end flanking sequences of beta(major) globin gene. Biochim. Biophys. Acta 2005, 1743, 101–114. [Google Scholar] [CrossRef]
- Akrivou, M.G.; Demertzidou, V.P.; Theodoroula, N.F.; Chatzopoulou, F.M.; Kyritsis, K.A.; Grigoriadis, N.; Zografos, A.L.; Vizirianakis, I.S. Uncovering the pharmacological response of novel sesquiterpene derivatives that differentially alter gene expression and modulate the cell cycle in cancer cells. Int. J. Oncol. 2018, 53, 2167–2179. [Google Scholar] [CrossRef] [Green Version]
- Tseligka, E.D.; Rova, A.; Amanatiadou, E.P.; Calabrese, G.; Tsibouklis, J.; Fatouros, D.G.; Vizirianakis, I.S. Pharmacological Development of Target-Specific Delocalized Lipophilic Cation-Functionalized Carboranes for Cancer Therapy. Pharm. Res. 2016, 33, 1945–1958. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |
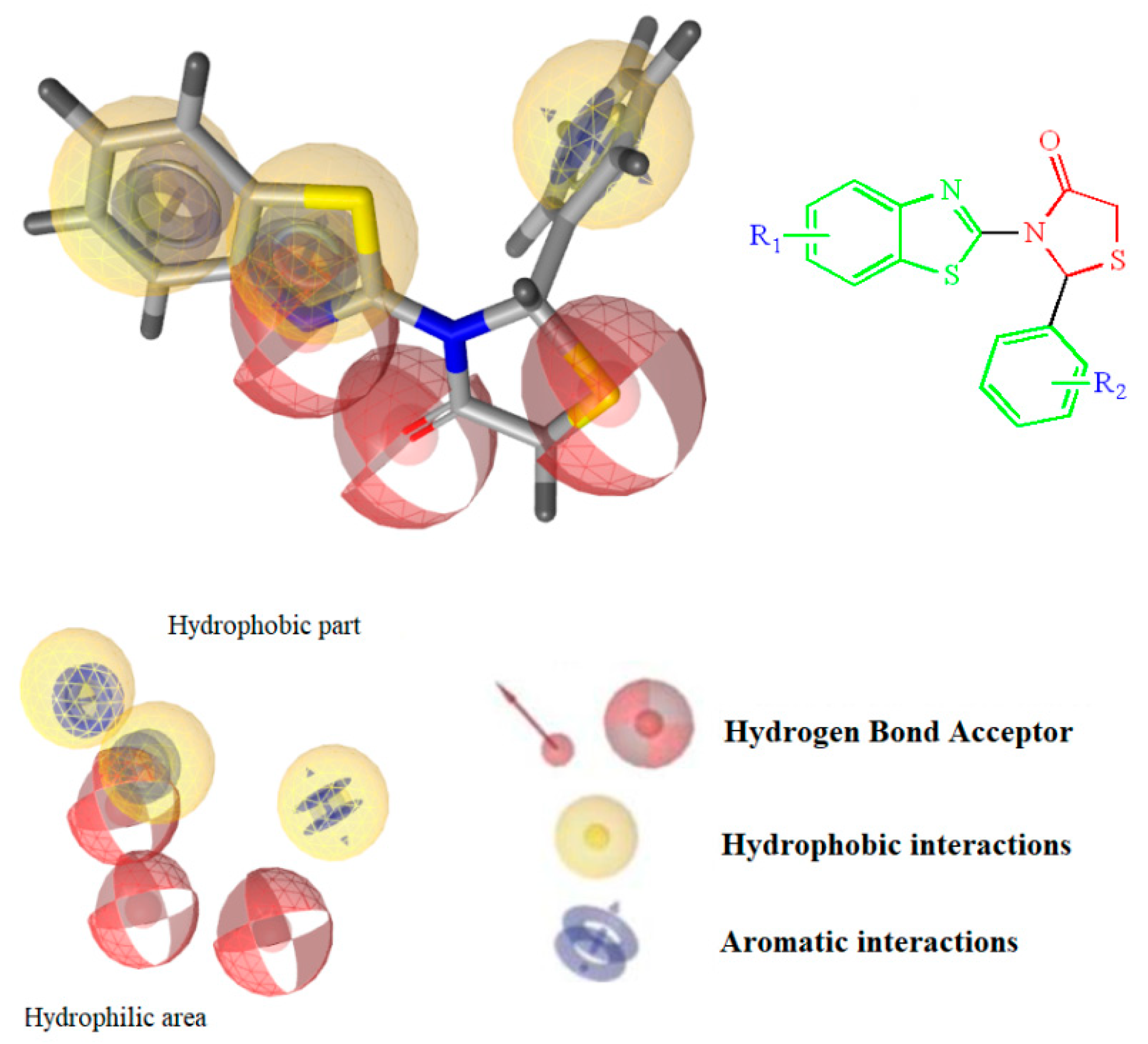






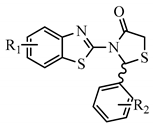
| N | R1 | R2 | Free Binding Energy (kcal/mol) S-(−) | Free Binding Energy (kcal/mol) R-(+) | Pa | Ν | R1 | R2 | Free Binding Energy (kcal/mol) -(−) | Free Binding Energy (kcal/mol) R-(+) | Pa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a1 (1) | 7-Cl | 2,6-di-F | −10.21 | −13.37 | 0.651 | i7 | 6-Br | 2,3-di-Cl | −3,25 | −4.07 | 0.352 |
| a2 (2) | 7-Cl | 2-F, 6-Cl | −11.42 | −14.10 | 0.704 | i8 | 6-Br | 2,4-di-Cl | −5,37 | −6.43 | 0.341 |
| b1 (3) | 6-F | 4-F | −10.95 | −13.21 | 0.472 | k1 | 6-CN | 3-Cl | −5.29 | −6.57 | 0.353 |
| b2 (4) | 6-F | 4-NO2 | −8.02 | −9.11 | 0.374 | k2 | 6-CN | 3-Br | −4.18 | −5.66 | 0.393 |
| b3 (5) | 6-F | 4-Cl | −8.15 | −9.02 | 0.398 | k3 | 6-CN | 3-F | −6.32 | −703 | 0.287 |
| b4 (6) | 6-F | 4-OCH3 | −7.14 | −8.70 | 0.360 | k4 | 6-CN | 2,6-di-Cl | −6.58 | −7.84 | 0.498 |
| b5 (7) | 6-F | 4-OH | −8.27 | −9.27 | 0.415 | k5 | 6-CN | 2,3-di-Cl | −6.45 | −7.16 | 0.447 |
| b6 | 6-F | 4-Br | −6.17 | −6.54 | 0.401 | k6 | 6-CN | 2,4-di-Cl | −6.93 | −7.18 | 0.366 |
| b7 | 6-F | 2,3-di-Cl | −6.53 | −6.71 | 0.353 | l1 | 6-CF3 | 2,6-di-Cl | −6.44 | −7.12 | 0.448 |
| b8 | 6-F | 2,4-di-Cl | −6.05 | −6.41 | 0.349 | l2 | 6-CF3 | 3-Cl | −5.41 | −6.27 | 0.332 |
| c1 (8) | 6-Cl | 4-F | −10.13 | −12.30 | 0.426 | l3 | 6-CF3 | 3-Br | −5.07 | −5.71 | 0.254 |
| c2 | 6-Cl | 2-Cl | −6.28 | −6.85 | 0.372 | l4 | 6-CF3 | 4-Br | −5.09 | −5.82 | 0.378 |
| c3 | 6-Cl | 3-Cl | −6.55 | −6.93 | 0.285 | l5 | 6-CF3 | 2,3-di-Cl | −4.05 | −4.70 | 0.336 |
| c4 | 6-Cl | 3-Br | −6.01 | −7.11 | 0.402 | l6 | 6-CF3 | 2,4-di-Cl | −6.41 | −6.91 | 0.332 |
| c5 (9) | 6-Cl | 4-OH | −11.08 | −13.63 | 0.492 | m1 | 6-Ad | 3-Cl | −5.12 | −6.74 | 0.273 |
| c6 | 6-Cl | 4-Br | −6.01 | −7.11 | 0.419 | m2 | 6-Ad | 3-Br | −5.96 | −6.89 | 0.280 |
| c7 | 6-Cl | 2,3-di-Cl | −6.25 | −7.59 | 0.411 | m3 | 6-Ad | 2,6-di-F | −6.25 | −7.04 | 0.428 |
| c8 | 6-Cl | 2,4-di-Cl | −5.92 | −6.42 | 0.419 | m4 | 6-Ad | 2,3-di-Cl | −7.14 | −8.05 | 0.289 |
| d1 (10) | 4-Cl | 4-F | −11.16 | −14.59 | 0.469 | m5 | 6-Ad | 2,4-di-Cl | −7.02 | −7.94 | 0.285 |
| d2 (11) | 4-Cl | 4-NO2 | −8.45 | −9.28 | 0.393 | m6 | 6-Ad | 4-F | −6.82 | −7.88 | 0.325 |
| d3 (12) | 4-Cl | 4-Cl | −8.73 | −10.26 | 0.480 | m7 | 6-Ad | 4-NO2 | −5.17 | −6.75 | 0.226 |
| d4 (13) | 4-Cl | 4-OCH3 | −9.11 | −11.69 | 0.378 | m8 | 6-Ad | 4-Cl | −6.79 | −7.55 | 0.319 |
| d5 (14) | 4-Cl | 4-OH | −10.58 | −13.72 | 0.436 | m9 | 6-Ad | 4-OCH3 | −6.49 | −7.28 | 0.295 |
| d6 | 4-Cl | 4-Br | −6.17 | −7.76 | 0.422 | m10 | 6-Ad | 4-OH | −4.18 | −5.94 | 0.333 |
| d7 | 4-Cl | 2,3-di-Cl | −6.63 | −7.15 | 0.424 | n1 | 4-CH3, 6-Ad | 3-Cl | −4.26 | −5.97 | 0.226 |
| d8 | 4-Cl | 2,4-di-Cl | −7.14 | −7.93 | 0.404 | n2 | 4-CH3, 6-Ad | 3-Br | −4.13 | −5.62 | 0.280 |
| e1 (15) | 4-OCH3 | 4-F | −9.93 | −11.51 | 0.353 | n3 | 4-CH3, 6-Ad | 2-F, 6-Cl | −7.03 | −8.12 | 0.507 |
| e2 (16) | 4-OCH3 | 4-NO2 | −10.05 | −11.58 | 0.374 | n4 | 4-CH3, 6-Ad | 2,3-di-Cl | −6.58 | −7.89 | 0.280 |
| e3 (17) | 4-OCH3 | 4-Cl | −9.92 | −11.23 | 0.347 | n5 | 4-CH3, 6-Ad | 2,4-di-Cl | −6.93 | −7.91 | 0.276 |
| e4 (18) | 4-OCH3 | 4-OCH3 | −8.76 | −10.97 | 0.380 | n6 | 4-CH3, 6-Ad | 4-F | −6.74 | −7.59 | 0.313 |
| e5 (19) | 4-OCH3 | 4-OH | −10.15 | −11.02 | 0.379 | n7 | 4-CH3, 6-Ad | 4-NO2 | −5.08 | −6.65 | 0.202 |
| e6 | 4-OCH3 | 4-Br | −6.58 | −7.46 | 0.329 | n8 | 4-CH3, 6-Ad | 4-Cl | −6.83 | −7.94 | 0.307 |
| e7 | 4-OCH3 | 2,3-di-Cl | −5.14 | −6.03 | 0.317 | n9 | 4-CH3, 6-Ad | 4-OCH3 | −6.37 | −7.17 | 0.287 |
| e8 | 4-OCH3 | 2,4-di-Cl | −5.79 | −6.48 | 0.322 | n10 | 4-CH3, 6-Ad | 4-OH | −4.16 | −5.93 | 0.321 |
| f1 (20) | 6-OCH3 | 4-F | −10.87 | −12.17 | 0.393 | o1 | 5,6-di-CH3 | 4-F | −6.95 | −7.56 | 0.433 |
| f2 (21) | 6-OCH3 | 4-NO2 | −7.14 | −8.96 | 0.346 | o2 | 5,6-di-CH3 | 4-NO2 | −5.18 | −6.05 | 0.328 |
| f3 (22) | 6-OCH3 | 4-Cl | −9.88 | −11.00 | 0.365 | o3 | 5,6-di-CH3 | 4-Cl | −5.21 | −6.17 | 0.432 |
| f4 (23) | 6-OCH3 | 4-OCH3 | −9.91 | −11.49 | 0.420 | o4 | 5,6-di-CH3 | 4-OCH3 | −3.28 | −4.56 | 0.327 |
| f5 | 6-OCH3 | 3-Br | −5.03 | −6.11 | 0.302 | o5 | 5,6-di-CH3 | 4-OH | −4.19 | −5.13 | 0.351 |
| f6 | 6-OCH3 | 4-Br | −5.17 | −6.13 | 0.367 | o6 | 5,6-di-CH3 | 4-Br | −3.05 | −4.27 | 0.355 |
| f7 | 6-OCH3 | 2,3-di-Cl | −5.61 | −6.92 | 0.303 | o7 | 5,6-di-CH3 | 2,3-di-Cl | −6.17 | −7.01 | 0.275 |
| f8 | 6-OCH3 | 2,4-di-Cl | −6.24 | −7.32 | 0.325 | o8 | 5,6-di-CH3 | 2,4-di-Cl | −6.14 | −6.86 | 0.286 |
| g1 (24) | 6-OCH2CH3 | 4-F | −6.97 | −8.35 | 0.373 | p1 | 6-NO2 | 4-F | −6.84 | −6.92 | 0.390 |
| g2 (25) | 6-OCH2CH3 | 4-NO2 | −7.14 | −9.02 | 0.339 | p2 | 6-NO2 | 4-NO2 | −5.11 | −6.01 | 0.225 |
| g3 (26) | 6-OCH2CH3 | 4-Cl | −9.84 | −11.35 | 0.376 | p3 | 6-NO2 | 4-Cl | −5.20 | −6.15 | 0.382 |
| g4 (27) | 6-OCH2CH3 | 4-OCH3 | −9.10 | −10.98 | 0.356 | p4 | 6-NO2 | 4-OCH3 | −3.15 | −4.42 | 0.348 |
| g5 (28) | 6-OCH2CH3 | 4-OH | −10.05 | −11.56 | 0.387 | p5 | 6-NO2 | 4-OH | −3.47 | −4.18 | 0.398 |
| g6 | 6-OCH2CH3 | 4-Br | −4.28 | −5.43 | 0.358 | p6 | 6-NO2 | 4-Br | −3.01 | −4.15 | 0.284 |
| g7 | 6-OCH2CH3 | 2,3-di-Cl | −5.84 | −6.47 | 0.286 | p7 | 6-NO2 | 2,3-di-Cl | −6.18 | −6.81 | 0.341 |
| g8 | 6-OCH2CH3 | 2,4-di-Cl | −5.33 | −6.21 | 0.320 | p8 | 6-NO2 | 2,4-di-Cl | −6.29 | −6.98 | 0.337 |
| h1 (29) | 6-OCF3 | 2,6-di-Cl | −9.87 | −11.03 | 0.388 | q1 | 4-CH3 | 4-F | −7.01 | −7.82 | 0.345 |
| h2 | 6-OCF3 | 2,6-di-F | −7.77 | −8.85 | 0.424 | q2 | 4-CH3 | 4-NO2 | −5.10 | −6.12 | 0.326 |
| h3 | 6-OCF3 | 3-Cl | −5.96 | −6.88 | 0.315 | q3 | 4-CH3 | 4-Cl | −5.03 | −5.85 | 0.335 |
| h4 (30) | 6-OCF3 | 2,3-di-Cl | −7.23 | −9.31 | 0.300 | q4 | 4-CH3 | 4-OCH3 | −4.85 | −5.16 | 0.290 |
| h5 | 6-OCF3 | 3-Br | −5.17 | −6.33 | 0.242 | q5 | 4-CH3 | 4-OH | −4.08 | −5.49 | 0.353 |
| h6 | 6-OCF3 | 4-F | −6.94 | −8.74 | 0.345 | q6 | 4-CH3 | 4-Br | −3.17 | −4.55 | 0.338 |
| h7 (31) | 6-OCF3 | 4-NO2 | −8.45 | −9.07 | 0.315 | q7 | 4-CH3 | 2,3-di-Cl | −6.14 | −6.80 | 0.390 |
| h8 (32) | 6-OCF3 | 4-Cl | −6.89 | −8.71 | 0.332 | q8 | 4-CH3 | 2,4-di-Cl | −6.33 | −6.86 | 0.382 |
| h9 | 6-OCF3 | 3-F | −5.22 | −6.19 | 0.330 | r1 | 6-CH3 | 4-F | −7.00 | −7.81 | 0.337 |
| h10 | 6-OCF3 | 4-OH | −7.97 | −8.51 | 0.442 | r2 | 6-CH3 | 4-NO2 | −5.02 | −5.93 | 0.315 |
| i1 | 6-Br | 4-F | −7.31 | −8.10 | 0.301 | r3 | 6-CH3 | 4-Cl | −5.01 | −5.83 | 0.327 |
| i2 | 6-Br | 4-NO2 | −6.88 | −7.95 | 0.326 | r4 | 6-CH3 | 4-OCH3 | −4.77 | −5.12 | 0.284 |
| i3 | 6-Br | 4-Cl | −7.19 | −8.02 | 0.393 | r5 | 6-CH3 | 4-OH | −4.05 | −5.40 | 0.345 |
| i4 | 6-Br | 4-OCH3 | −5.35 | −6.42 | 0.285 | r6 | 6-CH3 | 4-Br | −3.12 | −4.52 | 0.330 |
| i5 | 6-Br | 4-OH | −7.22 | −8.02 | 0.409 | r7 | 6-CH3 | 2,3-di-Cl | −6.13 | −6.78 | 0.377 |
| i6 | 6-Br | 4-Br | −5.11 | −5.84 | 0.405 | r8 | 6-CH3 | 2,4-di-Cl | −6.10 | −6.64 | 0.372 |
| Etravirine | −11.25 | Nevirapine | −11.95 | ||||||||
| N | Predicted Anti-HIV Activity (Pa) | Free Binding Energy (kcal/mol) S-(−) | Free Binding Energy (kcal/mol) R-(+) | H Bonds | Amino Acids | Hydrophobic Interactions | Pi Interact. | Halogen Interact. |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.651 | −10.21 | −13.37 | 1 | Lys101 | Thr139, Lys172, Gly190 | Pro95, Leu100, Lys103, Glu138, Val179, Tyr181 | Ile180 |
| 2 | 0.704 | −11.42 | −14.10 | 1 | Lys101 | Glu28, Gly99, Leu100, Lys103, Asn136, Tyr181, Ile382, Trp383 | Glu138, Ile135, Val179, Tyr319, Pro321 | Glu138 |
| 3 | 0.472 | −10.95 | −13.21 | 2 | Lys101 Trp229 | Val106, Gly190, Leu234 | Leu100, Lys103, Glu138, Val179, Tyr181, Tyr188, Trp229, Leu234 | - |
| 4 | 0.374 | −8.02 | −9.11 | 1 | Lys101 | Glu131, Thr132 | Leu100, Val106, Glu138 | - |
| 5 | 0.398 | −8.15 | −9.02 | - | - | Glu138, Tyr181, Tyr188, Gly190, Phe227, Tyr318 | Leu100, Lys101, Val106, Val179 | Leu234 Pro236 |
| 6 | 0.360 | −7.14 | −8.70 | 1 | Lys103 | Ser127, Glu131 | Leu100, Glu138, Val179, Tyr181 | - |
| 7 | 0.415 | −8.27 | −9.72 | - | - | Pro95, Lys101, Lys103, Tyre181, Gly190, Phe117, Trp229, His235, Pro236, Tyr318 | Leu100, Val106, Glu138, Val179, Tyr188, Leu234 | Glu138 |
| 8 | 0.426 | −10.13 | −12.30 | 2 | Lys101 Pro126 | Glu23, Ser127, Glu131, Thr132, Pro176, Ile178, Ile180 | Ile128, Arg172, Val179 | - |
| 9 | 0.492 | −11.08 | −14.63 | 2 | Lys101 Thr139 | Pro95, Lys103, Lys172, Tyr181, Leu234 | Leu100, Glu138, Val179 | - |
| 10 | 0.469 | −11.16 | −14.59 | 1 | Tyr318 | Ile180, Tur181, Gly190, Trp229 | Leu100, Lys103, Val106, Glu138, Val179, Tyr188, Leu234 | Lys101 His235 Pro236 |
| 11 | 0.393 | −8.45 | −9.28 | 1 | Lys101 | Lys103, Val106, Ile180 | Leu100, Val179, Tyr181 | - |
| 12 | 0.480 | −8.73 | −10.26 | 1 | Lys101 | Lys103, Ile180 | Leu100, Glu138, Val179 | |
| 13 | 0.378 | −9.11 | −11.69 | 2 | Lys101 Thr139 | Lys103, Lys172, Ile180 | Leu100, Glu138, Val179, Tyr181 | - |
| 14 | 0.436 | −10.58 | −13.72 | 3 | Lys101 Glu138 Thr139 | Val106, Leu234 | Leu100, Lys103, Val179, Tyr181 | - |
| 15 | 0.353 | −9.93 | −11.51 | 2 | Lys101 Trp229 | Pro95, Val106, Tyr188, Leu234 | Leu100, Lys103, Glu138, Val179, Tyr181 | - |
| 16 | 0.374 | −10.05 | −11.58 | 2 | Lys101 | Pro95, Val106, Tyr188 | Leu100, Glu138, Val179, Tyr181 | - |
| 17 | 0.347 | −9.92 | −11.23 | 1 | Lys101 | Pro95, Ile180, Tyr181, Gly190, Trp229, His235 | Leu100, Lys103, Glu138, Val179, Tyr188, Tyr318 | - |
| 18 | 0.380 | −8.76 | −10.97 | 2 | Glu138 Tyr318 | Pro95, Lys101, Lys102, Tyr181, Tyr188, Gly190, Trp229, His235 | Leu100, Val106, Glu138, Val179, Leu234 | - |
| 19 | 0.379 | −10.15 | −11.02 | 1 | Lys101 | Lys103, Ile180, Tyr181, Gly190, Trp229, Pro236, Tyr318, His235 | Leu100, Val106, Glu138, Val179, Tyr188, Leu234 | - |
| 20 | 0.393 | 10.87 | −12.17 | 2 | Lys101 Thr139 | Pro95, Leu100, Lys103, Lys172, Ile180, Tyr188 | Glu138, Val179, Tyr181, Trp229 | - |
| 21 | 0.346 | −7.14 | −8.96 | 1 | Lys101 | Pro95, Ile180, Tyr188 | Leu100, Glu138, Val179 | - |
| 22 | 0.365 | −9.88 | −11.00 | 2 | Lys101 Thr132 | Pro95, Lys172, Ile180, Tyr188 | Leu100, Glu138, Val179 | - |
| 23 | 0.420 | −9.91 | −11.49 | 2 | Lys101 Tyr181 | Lys102, Lys103, Ile180, Tyr188, Phe225, Pro234 | Leu100, Val106, Val179, Trp227, Leu232, Tyr316 | - |
| 24 | 0.373 | −6.97 | −8.35 | 1 | Lys101 | Pro95, Lys103, Ile180, Tyr181 | Leu100, Glu138, Val179 | - |
| 25 | 0.339 | −7.14 | −9.02 | 1 | Lys101 | Pro95, Gly99, Thr139, Ile180, Tyr181 | Leu100, Glu138, Val179 | - |
| 26 | 0.376 | −9.84 | −11.35 | 1 | Lys101 | Pro95, Gly99, Lys103, Thr139, Lys172, Ile180, Tyr181 | Leu100, Glu138, Val179, Trp229, Leu234 | - |
| 27 | 0.356 | −9.10 | −10.98 | 1 | Lys101 | Pro95, Pro97, Lys103, Thr139, Lys172, Ile180, Tyr181 | Leu100, Glu138, Val179, Trp229, Leu234 | - |
| 28 | 0.387 | −10.05 | −11.56 | 2 | Lys101 Thr139 | Pro95, Pro97, Lys103, Lys172, Ile180 | Leu100, Glu138, Val179, Trp229, Leu234 | - |
| 29 | 0.388 | −9.87 | −11.03 | 1 | Lys101 | Pro95, Tyr181, Gly190, Phe227, Pro236, Tyr318 | Leu100, Lys103, Val106, Val179, Tyr188, Leu234 | Glu138 Gly99 |
| 30 | 0.300 | −7.23 | −9.31 | 1 | Lys101 | Lys101, Tyr181, Gly190, His235 | Leu100, Val179, Tyr188 | - |
| 31 | 0.315 | −8.45 | −9.07 | 1 | Lys101 | Pro95, Lys103, Lys172, Leu234 | Leu100, Glu138, Val179, | - |
| 32 | 0.332 | −6.89 | −8.71 | 2 | Lys103 Thr132 | Pro95, Tyr181 | Glu138, Val179, Pro321 | - |
| Nevirapine | −11.95 | 2 | Lys101, Glu138 | Glu28, Gly99, Leu100, Lys101, Lys103, Val179, Ile382 | Lys101, Glu138, Ile135, Tyr319, Trp383 | - | ||
| Etravirine | −11.25 | 2 | Lys101, Glu138 | Lys102, Val108, Ty188, Pro225, Phe227, Pro236 | Pro95, Leu100, Lys103, Val106, Val179, Tyr181, Thr229, Leu234 | - | ||
| Έ. | Predicted LD50 | Predicted Toxicity Class | Hepatotoxicity | Carcinogenicity | Immunotoxicity | Mutagenicity | Cytotoxicity |
|---|---|---|---|---|---|---|---|
| 1 | 500 mg/kg | 4 | Inactive 0.56 | Inactive 0.57 | Inactive 0.94 | Inactive 0.67 | Inactive 0.76 |
| 2 | 500 mg/kg | 4 | Inactive 0.56 | Inactive 0.57 | Inactive 0.95 | Inactive 0.67 | Inactive 0.76 |
| 3 | 500 mg/kg | 4 | Inactive 0.57 | Inactive 0.55 | Inactive 0.99 | Inactive 0.66 | Inactive 0.80 |
| 4 | 1000 mg/kg | 4 | Inactive 0.55 | Inactive 0.68 | Inactive 0.99 | Inactive 0.66 | Inactive 0.73 |
| 5 | 500 mg/kg | 4 | Inactive 0.57 | Inactive 0.55 | Inactive 0.99 | Inactive 0.67 | Inactive 0.81 |
| 6 | 500 mg/kg | 4 | Inactive 0.60 | Inactive 0.58 | Inactive 0.98 | Inactive 0.65 | Inactive 0.66 |
| 7 | 500 mg/kg | 4 | Inactive 0.63 | Inactive 0.59 | Inactive 0.99 | Inactive 0.66 | Inactive 0.70 |
| 8 | 500 mg/kg | 4 | Inactive 0.57 | Inactive 0.55 | Inactive 0.99 | Inactive 0.67 | Inactive 0.81 |
| 9 | 500 mg/kg | 4 | Inactive 0.60 | Inactive 0.58 | Inactive 0.99 | Inactive 0.68 | Inactive 0.68 |
| 10 | 500 mg/kg | 4 | Inactive 0.57 | Inactive 0.55 | Inactive 0.99 | Inactive 0.67 | Inactive 0.81 |
| 11 | 1000 mg/kg | 4 | Inactive 0.52 | Inactive 0.65 | Inactive 0.99 | Active 0.91 | Inactive 0.74 |
| 12 | 500 mg/kg | 4 | Inactive 0.54 | Inactive 0.55 | Inactive 0.99 | Inactive 0.68 | Inactive 0.82 |
| 13 | 500 mg/kg | 4 | Inactive 0.57 | Inactive 0.58 | Inactive 0.99 | Inactive 0.66 | Inactive 0.64 |
| 14 | 500 mg/kg | 4 | Inactive 0.60 | Inactive 0.58 | Inactive 0.99 | Inactive 0.68 | Inactive 0.68 |
| 15 | 500 mg/kg | 4 | Inactive 0.59 | Inactive 0.60 | Inactive 0.98 | Inactive 0.66 | Inactive 0.66 |
| 16 | 1000 mg/kg | 4 | Inactive 0.52 | Inactive 0.65 | Inactive 0.99 | Inactive 0.66 | Inactive 0.74 |
| 17 | 500 mg/kg | 4 | Inactive 0.56 | Inactive 0.60 | Inactive 0.98 | Inactive 0.67 | Inactive 0.65 |
| 18 | 500 mg/kg | 4 | Inactive 0.50 | Inactive 0.62 | Inactive 0.98 | Inactive 0.61 | Inactive 0.63 |
| 19 | 500 mg/kg | 4 | Inactive 0.52 | Inactive 0.64 | Inactive 0.99 | Inactive 0.60 | Inactive 0.63 |
| 20 | 500 mg/kg | 4 | Inactive 0.59 | Inactive 0.60 | Inactive 0.98 | Inactive 0.66 | Inactive 0.66 |
| 21 | 1000 mg/kg | 4 | Inactive 0.52 | Inactive 0.65 | Inactive 0.99 | Inactive 0.66 | Inactive 0.74 |
| 22 | 500 mg/kg | 4 | Inactive 0.56 | Inactive 0.60 | Inactive 0.98 | Inactive 0.67 | Inactive 0.65 |
| 23 | 500 mg/kg | 4 | Inactive 0.50 | Inactive 0.62 | Inactive 0.98 | Inactive 0.61 | Inactive 0.63 |
| 24 | 500 mg/kg | 4 | Inactive 0.52 | Inactive 0.57 | Inactive 0.96 | Inactive 0.64 | Inactive 0.62 |
| 25 | 1000 mg/kg | 4 | Inactive 0.53 | Inactive 0.63 | Inactive 0.97 | Inactive 0.66 | Inactive 0.62 |
| 26 | 500 mg/kg | 4 | Inactive 0.52 | Inactive 0.56 | Inactive 0.98 | Inactive 0.65 | Inactive 0.59 |
| 27 | 500 mg/kg | 4 | Inactive 0.53 | Inactive 0.62 | Inactive 0.97 | Inactive 0.61 | Inactive 0.61 |
| 28 | 500 mg/kg | 4 | Inactive 0.50 | Inactive 0.62 | Inactive 0.99 | Inactive 0.63 | Inactive 0.63 |
| 29 | 500 mg/kg | 4 | Inactive 0.61 | Inactive 0.56 | Inactive 0.91 | Inactive 0.66 | Inactive 0.64 |
| 30 | 500 mg/kg | 4 | Inactive 0.61 | Inactive 0.56 | Inactive 0.91 | Inactive 0.66 | Inactive 0.64 |
| 31 | 1000 mg/kg | 4 | Inactive 0.56 | Inactive 0.64 | Inactive 0.81 | Inactive 0.66 | Inactive 0.64 |
| 32 | 500 mg/kg | 4 | Inactive 0.61 | Inactive 0.56 | Inactive 0.97 | Inactive 0.66 | Inactive 0.65 |
| N | Inhibition % (4 μM) | IC50 (μM) | N | Inhibition % (4 μM) | IC50 (μM) | N | Inhibition % (4 μM) | IC50 (μM) |
|---|---|---|---|---|---|---|---|---|
| 1 | 70 | 0.21 | 12 | 50 | 4 | 23 | 73 | 2.39 |
| 2 | 72 | 0.0047 | 13 | 65 | 1.99 | 24 | 36 | >4 |
| 3 | 72 | 0.15 | 14 | 83 | 0.01 | 25 | 44 | >4 |
| 4 | 54 | 3.8 | 15 | 69 | 2.08 | 26 | 73 | 2.48 |
| 5 | 44 | >4 | 16 | 68 | 2.20 | 27 | 76 | 1.93 |
| 6 | 35 | >4 | 17 | 67 | 2.60 | 28 | 62 | 2.00 |
| 7 | 50 | 4 | 18 | 72 | 2.69 | 29 | 49 | 4.28 |
| 8 | 83 | 0.26 | 19 | 71 | 2.62 | 30 | 46 | >4 |
| 9 | 56 | 0.001 | 20 | 55 | 1.51 | 31 | 46 | >4 |
| 10 | 63 | 0.001 | 21 | 50 | 4 | 32 | 46 | >4 |
| 11 | 40 | >4 | 22 | 72 | 2.76 | Nevirapine | 0.3 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrou, A.; Eleftheriou, P.; Geronikaki, A.; Akrivou, M.G.; Vizirianakis, I. Novel Thiazolidin-4-ones as Potential Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase. Molecules 2019, 24, 3821. https://doi.org/10.3390/molecules24213821
Petrou A, Eleftheriou P, Geronikaki A, Akrivou MG, Vizirianakis I. Novel Thiazolidin-4-ones as Potential Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase. Molecules. 2019; 24(21):3821. https://doi.org/10.3390/molecules24213821
Chicago/Turabian StylePetrou, Anthi, Phaedra Eleftheriou, Athina Geronikaki, Melpomeni G. Akrivou, and Ioannis Vizirianakis. 2019. "Novel Thiazolidin-4-ones as Potential Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase" Molecules 24, no. 21: 3821. https://doi.org/10.3390/molecules24213821
APA StylePetrou, A., Eleftheriou, P., Geronikaki, A., Akrivou, M. G., & Vizirianakis, I. (2019). Novel Thiazolidin-4-ones as Potential Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase. Molecules, 24(21), 3821. https://doi.org/10.3390/molecules24213821








