Synthesis of Novel Farnesoid X Receptor Agonists and Validation of Their Efficacy in Activating Differentiation of Mouse Bone Marrow-Derived Mesenchymal Stem Cells into Osteoblasts
Abstract
:1. Introduction
2. Results
2.1. Synthesis of Novel FXR Agonists
2.2. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) Binding Assay
2.3. Specificity of FXR Agonists as Determined by Luciferase Reporter Assay
2.4. Viability of ST-2 MSCs
2.5. Expression of FXR and FXR-Target Genes during Osteoblast Differentiation of ST-2 MSCs
2.6. Enhancement of ALP Activity by FXR Agonists during Osteoblast Differentiation of ST-2 MSCs
2.7. Elevation of the Expression of Osteogenic Genes by FXR Agonists in ST-2 MSCs
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of FXR Agonists
4.3. TR-FRET Binding Assay
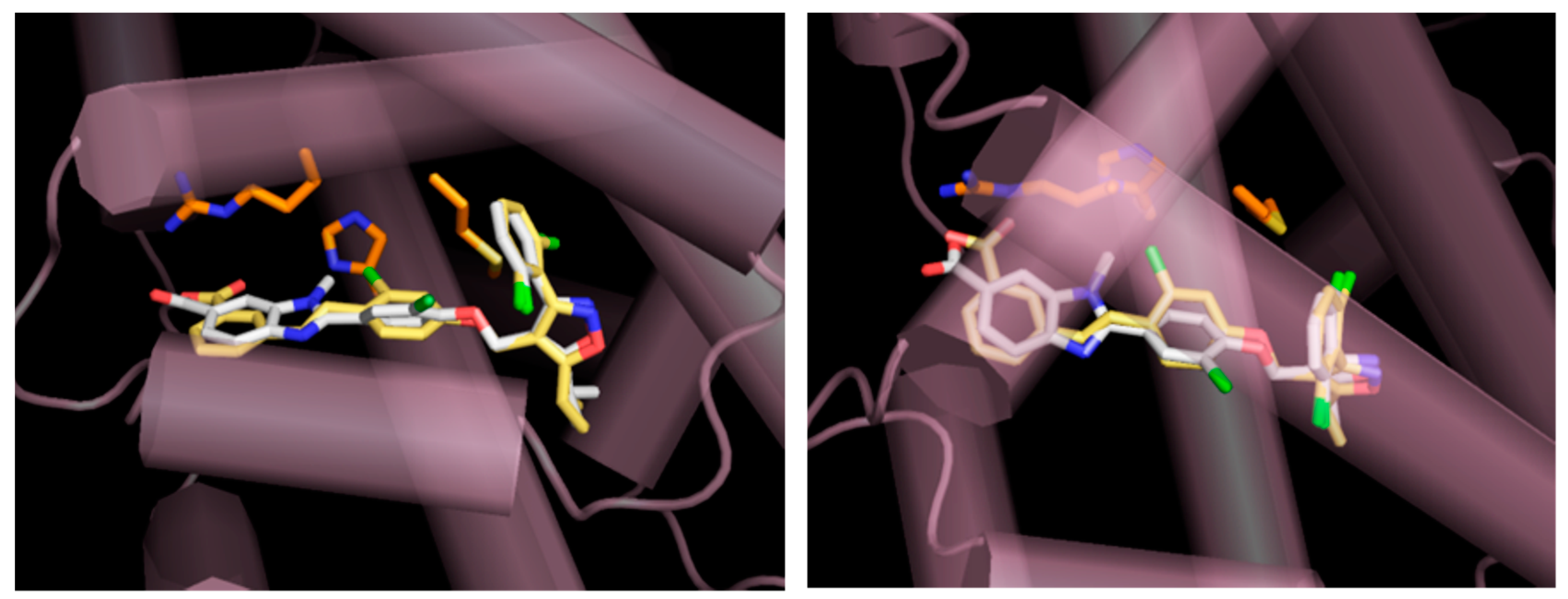
4.4. Modeling of FXR Complex with GW4064 and Novel FXR Agonist 3
4.5. Luciferase Reporter Assay
4.6. Cell Culture
4.7. Cell Toxicity Assay
4.8. RNA Extraction and Quantitative PCR
4.9. ALP Staining
4.10. ALP Activity Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, C.; Cho, K.; Huang, Y.; Lyons, J.P.; Zhou, X.; Sinha, K.; McCrea, P.D.; de Crombrugghe, B. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc. Natl. Acad. Sci. USA 2008, 105, 6936–6941. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Nishikawa, S.; Ikuta, K.; Yamamura, F.; Naito, M.; Takahashi, K.; Nishikawa, S. B cell ontogeny in murine embryo studied by a culture system with the monolayer of a stromal cell clone, ST2: B cell progenitor develops first in the embryonal body rather than in the yolk sac. EMBO J. 1988, 7, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jahagirdar, B.N.; Reinhardt, R.L.; Schwartz, R.E.; Keene, C.D.; Ortiz-Gonzalez, X.R.; Reyes, M.; Lenvik, T.; Lund, T.; Blackstad, M.; et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002, 418, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef]
- Spees, J.L.; Lee, R.H.; Gregory, C.A. Mechanisms of mesenchymal stem/stromal cell function. Stem Cell Res. Ther. 2016, 7, 125. [Google Scholar] [CrossRef]
- Hadjidakis, D.J.; Androulakis, I.I. Bone remodeling. Ann. N. Y. Acad. Sci. 2006, 1092, 385–396. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Lian, J.B.; Javed, A.; Zaidi, S.K.; Lengner, C.; Montecino, M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Regulatory controls for osteoblast growth and differentiation: Role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 2004, 14, 1–41. [Google Scholar] [CrossRef]
- Komori, T. Regulation of skeletal development by the Runx family of transcription factors. J. Cell. Biochem. 2005, 95, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Harada, H.; Tagashira, S.; Fujiwara, M.; Ogawa, S.; Katsumata, T.; Yamaguchi, A.; Komori, T.; Nakatsuka, M. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J. Biol. Chem. 1999, 274, 6972–6978. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Takarada, T.; Hinoi, E.; Nakazato, R.; Ochi, H.; Xu, C.; Tsuchikane, A.; Takeda, S.; Karsenty, G.; Abe, T.; Kiyonari, H.; et al. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J. Bone Miner. Res. 2013, 28, 2064–2069. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Goode, E.; Chen, J.; Oro, A.E.; Bradley, D.J.; Perlmann, T.; Noonan, D.J.; Burka, L.T.; McMorris, T.; Lamph, W.W.; et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 1995, 81, 687–693. [Google Scholar] [CrossRef]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a nuclear receptor for bile acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.T.; Makishima, M.; Repa, J.J.; Schoonjans, K.; Kerr, T.A.; Auwerx, J.; Mangelsdorf, D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 2000, 6, 507–515. [Google Scholar] [CrossRef]
- Chiang, J.Y. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr. Rev. 2002, 23, 443–463. [Google Scholar] [CrossRef]
- Moore, D.D.; Kato, S.; Xie, W.; Mangelsdorf, D.J.; Schmidt, D.R.; Xiao, R.; Kliewer, S.A. International Union of Pharmacology. LXII. The NR1H and NR1I receptors: Constitutive androstane receptor, pregnene X receptor, farnesoid X receptor α, farnesoid X receptor β, liver X receptor α, liver X receptor β, and vitamin D receptor. Pharmacol. Rev. 2006, 58, 742–759. [Google Scholar] [CrossRef]
- Pellicciari, R.; Costantino, G.; Fiorucci, S. Farnesoid X receptor: From structure to potential clinical applications. J. Med. Chem. 2005, 48, 5383–5403. [Google Scholar] [CrossRef]
- Fiorucci, S.; Rizzo, G.; Donini, A.; Distrutti, E.; Santucci, L. Targeting farnesoid X receptor for liver and metabolic disorders. Trends Mol. Med. 2007, 13, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.Y.; Lee, H.; Hubbert, M.L.; Edwards, P.A.; Zhang, Y. FXR, a multipurpose nuclear receptor. Trends Biochem. Sci. 2006, 31, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Urizar, N.L.; Dowhan, D.H.; Moore, D.D. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J. Biol. Chem. 2000, 275, 39313–39317. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.C.; Yang, J.S.; Lu, H.F.; Ip, S.W.; Lo, C.; Wu, C.C.; Lin, J.P.; Tang, N.Y.; Chung, J.G.; Chou, M.J.; et al. Quercetin-mediated cell cycle arrest and apoptosis involving activation of a caspase cascade through the mitochondrial pathway in human breast cancer MCF-7 cells. Arch. Pharm. Res. 2010, 33, 1181–1191. [Google Scholar] [CrossRef]
- Id Boufker, H.; Lagneaux, L.; Fayyad-Kazan, H.; Badran, B.; Najar, M.; Wiedig, M.; Ghanem, G.; Laurent, G.; Body, J.J.; Journe, F. Role of farnesoid X receptor (FXR) in the process of differentiation of bone marrow stromal cells into osteoblasts. Bone 2011, 49, 1219–1231. [Google Scholar] [CrossRef]
- Cho, S.W.; An, J.H.; Park, H.; Yang, J.Y.; Choi, H.J.; Kim, S.W.; Park, Y.J.; Kim, S.Y.; Yim, M.; Baek, W.Y.; et al. Positive regulation of osteogenesis by bile acid through FXR. J. Bone Miner. Res. 2013, 28, 2109–2121. [Google Scholar] [CrossRef]
- Jeong, B.C.; Lee, Y.S.; Bae, I.H.; Lee, C.H.; Shin, H.I.; Ha, H.J.; Franceschi, R.T.; Choi, H.S.; Koh, J.T. The orphan nuclear receptor SHP is a positive regulator of osteoblastic bone formation. J. Bone Miner. Res. 2010, 25, 262–274. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Evans, R.M.; Roecker, A.J.; Hughes, R.; Downes, M.; Pfefferkorn, J.A. Discovery and optimization of non-steroidal FXR agonists from natural product-like libraries. Org. Biomol. Chem. 2003, 1, 908–920. [Google Scholar] [CrossRef]
- Markham, A.; Keam, S.J. Obeticholic acid: First global approval. Drugs 2016, 76, 1221–1226. [Google Scholar] [CrossRef]
- Armstrong, L.E.; Guo, G.L. Role of FXR in liver inflammation during nonalcoholic steatohepatitis. Curr. Pharmacol. Rep. 2017, 3, 92–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gege, C.; Hambruch, E.; Hambruch, N.; Kinzel, O.; Kremoser, C. Nonsteroidal FXR Ligands: Current status and clinical applications. Handb. Exp. Pharmacol. 2019, 256, 167–205. [Google Scholar] [CrossRef] [PubMed]
- Tully, D.C.; Rucker, P.V.; Chianelli, D.; Williams, J.; Vidal, A.; Alper, P.B.; Mutnick, D.; Bursulaya, B.; Schmeits, J.; Wu, X.; et al. Discovery of Tropifexor (LJN452), a highly potent non-bile acid FXR agonist for the treatment of cholestatic liver diseases and nonalcoholic steatohepatitis (NASH). J. Med. Chem. 2017, 60, 9960–9973. [Google Scholar] [CrossRef] [PubMed]
- Maloney, P.R.; Parks, D.J.; Haffner, C.D.; Fivush, A.M.; Chandra, G.; Plunket, K.D.; Creech, K.L.; Moore, L.B.; Wilson, J.G.; Lewis, M.C.; et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 2000, 43, 2971–2974. [Google Scholar] [CrossRef] [PubMed]
- Akwabi-Ameyaw, A.; Bass, J.Y.; Caldwell, R.D.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Jones, S.A.; Kaldor, I.; Liu, Y.; et al. Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064. Bioorg. Med. Chem. Lett. 2008, 18, 4339–4343. [Google Scholar] [CrossRef]
- Bass, J.Y.; Caldwell, R.D.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Madauss, K.P.; Marr, H.B.; McFadyen, R.B.; Miller, A.B.; et al. Substituted isoxazole analogs of farnesoid X receptor (FXR) agonist GW4064. Bioorg. Med. Chem. Lett. 2009, 19, 2969–2973. [Google Scholar] [CrossRef]
- Akwabi-Ameyaw, A.; Bass, J.Y.; Caldwell, R.D.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Madauss, K.P.; Marr, H.B.; McFadyen, R.B.; et al. FXR agonist activity of conformationally constrained analogs of GW 4064. Bioorg. Med. Chem. Lett. 2009, 19, 4733–4739. [Google Scholar] [CrossRef]
- Bass, J.Y.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Madauss, K.P.; Marr, H.B.; McFadyen, R.B.; Miller, A.B.; Mills, W.Y.; et al. Conformationally constrained farnesoid X receptor (FXR) agonists: Heteroaryl replacements of the naphthalene. Bioorg. Med. Chem. Lett. 2011, 21, 1206–1213. [Google Scholar] [CrossRef]
- Akwabi-Ameyaw, A.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Madauss, K.P.; Marr, H.B.; Miller, A.B.; Navas, F., 3rd; Parks, D.J.; et al. Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene. Bioorg. Med. Chem. Lett. 2011, 21, 6154–6160. [Google Scholar] [CrossRef]
- Smalley, T.L., Jr.; Boggs, S.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Kaldor, I.; Parks, D.J. Novel heterocyclic scaffolds of GW4064 as farnesoid X receptor agonists. Bioorg. Med. Chem. Lett. 2015, 25, 280–284. [Google Scholar] [CrossRef]
- Genin, M.J.; Bueno, A.B.; Agejas Francisco, J.; Manninen, P.R.; Bocchinfuso, W.P.; Montrose-Rafizadeh, C.; Cannady, E.A.; Jones, T.M.; Stille, J.R.; Raddad, E.; et al. Discovery of 6-(4-{[5-cyclopropyl-3-(2,6-dichlorophenyl)isoxazol-4-yl]methoxy}piperidin-1-yl)-1-methyl-1H-indole-3-cARBOXYLIC acid: A novel FXR agonist for the treatment of dyslipidemia. J. Med. Chem. 2015, 58, 9768–9772. [Google Scholar] [CrossRef] [PubMed]
- Kinzel, O.; Steeneck, C.; Schluter, T.; Schulz, A.; Gege, C.; Hahn, U.; Hambruch, E.; Hornberger, M.; Spalwisz, A.; Frick, K.; et al. Novel substituted isoxazole FXR agonists with cyclopropyl, hydroxycyclobutyl and hydroxyazetidinyl linkers: Understanding and improving key determinants of pharmacological properties. Bioorg. Med. Chem. Lett. 2016, 26, 3746–3753. [Google Scholar] [CrossRef] [PubMed]
- Sepe, V.; Marchiano, S.; Finamore, C.; Baronissi, G.; Di Leva, F.S.; Carino, A.; Biagioli, M.; Fiorucci, C.; Cassiano, C.; Monti, M.C.; et al. Novel isoxazole derivatives with potent FXR agonistic activity prevent acetaminophen-induced liver injury. ACS Med. Chem. Lett. 2019, 10, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Kainuma, M.; Kasuga, J.; Hosoda, S.; Wakabayashi, K.; Tanatani, A.; Nagasawa, K.; Miyachi, H.; Makishima, M.; Hashimoto, Y. Diphenylmethane skeleton as a multi-template for nuclear receptor ligands: Preparation of FXR and PPAR ligands. Bioorg. Med. Chem. Lett. 2006, 16, 3213–3218. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, S.; Hashimoto, Y. Structural development studies of nuclear receptor ligands. Pure Appl. Chem. 2007, 79, 615–626. [Google Scholar] [CrossRef]
- Deaton, D.N.; Navas, F.I.; Spearing, P.K. Farnesoid X Receptor Agonists. Patent WO 2008157270, 13 June 2008. [Google Scholar]
- Flesch, D.; Gabler, M.; Lill, A.; Gomez, R.C.; Steri, R.; Schneider, G.; Stark, H.; Schubert-Zsilavecz, M.; Merk, D. Fragmentation of GW4064 led to a highly potent partial farnesoid X receptor agonist with improved drug-like properties. Bioorg. Med. Chem. 2015, 23, 3490–3498. [Google Scholar] [CrossRef]
- Merk, D.; Lamers, C.; Ahmad, K.; Carrasco Gomez, R.; Schneider, G.; Steinhilber, D.; Schubert-Zsilavecz, M. Extending the structure-activity relationship of anthranilic acid derivatives as farnesoid X receptor modulators: Development of a highly potent partial farnesoid X receptor agonist. J. Med. Chem. 2014, 57, 8035–8055. [Google Scholar] [CrossRef]
- Flesch, D.; Cheung, S.Y.; Schmidt, J.; Gabler, M.; Heitel, P.; Kramer, J.; Kaiser, A.; Hartmann, M.; Lindner, M.; Luddens-Damgen, K.; et al. Nonacidic farnesoid X receptor modulators. J. Med. Chem. 2017, 60, 7199–7205. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Z.; Zhou, J.; Guo, Y.; Wang, G.; Hao, H.; Xu, X. A novel intestinal-restricted FXR agonist. Bioorg. Med. Chem. Lett. 2017, 27, 3386–3390. [Google Scholar] [CrossRef]
- Richter, H.G.; Benson, G.M.; Bleicher, K.H.; Blum, D.; Chaput, E.; Clemann, N.; Feng, S.; Gardes, C.; Grether, U.; Hartman, P.; et al. Optimization of a novel class of benzimidazole-based farnesoid X receptor (FXR) agonists to improve physicochemical and ADME properties. Bioorg. Med. Chem. Lett. 2011, 21, 1134–1140. [Google Scholar] [CrossRef]
- Teno, N.; Iguchi, Y.; Yamashita, Y.; Mori, N.; Une, M.; Nishimaki-Mogami, T.; Gohda, K. Discovery and optimization of benzimidazole derivatives as a novel chemotype of farnesoid X receptor (FXR) antagonists. Bioorg. Med. Chem. 2017, 25, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Teno, N.; Yamashita, Y.; Iguchi, Y.; Fujimori, K.; Une, M.; Nishimaki-Mogami, T.; Hiramoto, T.; Gohda, K. Nonacidic chemotype possessing N-acylated piperidine moiety as potent farnesoid X Receptor (FXR) antagonists. ACS Med. Chem. Lett. 2018, 9, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Teno, N.; Yamashita, Y.; Masuda, A.; Iguchi, Y.; Oda, K.; Fujimori, K.; Hiramoto, T.; Nishimaki-Mogami, T.; Une, M.; Gohda, K. Identification of potent farnesoid X receptor (FXR) antagonist showing favorable PK profile and distribution toward target tissues: Comprehensive understanding of structure-activity relationship of FXR antagonists. Bioorg. Med. Chem. 2019, 27, 2220–2227. [Google Scholar] [CrossRef] [PubMed]
- Luan, Z.L.; Huo, X.K.; Dong, P.P.; Tian, X.G.; Sun, C.P.; Lv, X.; Feng, L.; Ning, J.; Wang, C.; Zhang, B.J.; et al. Highly potent non-steroidal FXR agonists protostane-type triterpenoids: Structure-activity relationship and mechanism. Eur. J. Med. Chem. 2019, 182, 111652. [Google Scholar] [CrossRef]
- Giancristofaro, A.; Barbosa, A.J.M.; Ammazzalorso, A.; Amoia, P.; De Filippis, B.; Fantacuzzi, M.; Giampietro, L.; Maccallini, C.; Amoroso, R. Discovery of new FXR agonists based on 6-ECDCA binding properties by virtual screening and molecular docking. MedChemComm 2018, 9, 1630–1638. [Google Scholar] [CrossRef]
- Massafra, V.; Pellicciari, R.; Gioiello, A.; van Mil, S.W.C. Progress and challenges of selective Farnesoid X Receptor modulation. Pharmacol. Ther. 2018, 191, 162–177. [Google Scholar] [CrossRef]
- Keri, R.S.; Hiremathad, A.; Budagumpi, S.; Nagaraja, B.M. Comprehensive review in current developments of benzimidazole-based medicinal chemistry. Chem. Biol. Drug Des. 2015, 86, 19–65. [Google Scholar] [CrossRef]
- Largeron, M.; Nguyen, K.M.H. Recent advances in the synthesis of benzimidazole derivatives from the oxidative coupling of primary amines. Synthesis 2018, 50, 241–253. [Google Scholar] [CrossRef]
- Yadav, G.; Ganguly, S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: A mini-review. Eur. J. Med. Chem. 2015, 97, 419–443. [Google Scholar] [CrossRef]
- Alasmary, F.A.; Snelling, A.M.; Zain, M.E.; Alafeefy, A.M.; Awaad, A.S.; Karodia, N. Synthesis and evaluation of selected benzimidazole derivatives as potential antimicrobial agents. Molecules 2015, 20, 15206–15223. [Google Scholar] [CrossRef]
- Keri, R.S.; Chand, K.; Budagumpi, S.; Balappa Somappa, S.; Patil, S.A.; Nagaraja, B.M. An overview of benzo[b]thiophene-based medicinal chemistry. Eur. J. Med. Chem. 2017, 138, 1002–1033. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.Z.; Devarakonda, S.; Harp, J.M.; Han, Q.; Pellicciari, R.; Willson, T.M.; Khorasanizadeh, S.; Rastinejad, F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell 2003, 11, 1093–1100. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell. Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, Y.; Youn, M.Y.; Inoue, K.; Takada, I.; Kouzmenko, A.; Kato, S. Nuclear receptors in bone physiology and diseases. Physiol. Rev. 2013, 93, 481–523. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast-osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef]
- Zheng, T.; Kang, J.H.; Sim, J.S.; Kim, J.W.; Koh, J.T.; Shin, C.S.; Lim, H.; Yim, M. The farnesoid X receptor negatively regulates osteoclastogenesis in bone remodeling and pathological bone loss. Oncotarget 2017, 8, 76558–76573. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.H.; Javed, A.; Kim, H.J.; Shin, H.I.; Gutierrez, S.; Choi, J.Y.; Rosen, V.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S.; et al. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor beta1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J. Cell. Biochem. 1999, 73, 114–125. [Google Scholar] [CrossRef]
- Lee, K.S.; Kim, H.J.; Li, Q.L.; Chi, X.Z.; Ueta, C.; Komori, T.; Wozney, J.M.; Kim, E.G.; Choi, J.Y.; Ryoo, H.M.; et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 2000, 20, 8783–8792. [Google Scholar] [CrossRef] [Green Version]
- Bruderer, M.; Richards, R.G.; Alini, M.; Stoddart, M.J. Role and regulation of RUNX2 in osteogenesis. Eur. Cell. Mater. 2014, 28, 269–286. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Tare, R.S.; Lee, S.H.; Mandeville, M.; Morasso, M.I.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J. Biol. Chem. 2006, 281, 40515–40526. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.H.; Kim, Y.J.; Kim, H.J.; Park, H.D.; Kang, A.R.; Kyung, H.M.; Sung, J.H.; Wozney, J.M.; Kim, H.J.; Ryoo, H.M. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 2003, 278, 34387–34394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iguchi, Y.; Yamaguchi, M.; Sato, H.; Kihira, K.; Nishimaki-Mogami, T.; Une, M. Bile alcohols function as the ligands of membrane-type bile acid-activated G protein-coupled receptor. J. Lipid Res. 2010, 51, 1432–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |






| FXR Agonist | EC50 (nM) | Maximum Activity (%) |
|---|---|---|
| GW4064 | 96.2 ± 62.7 | 135.8 ± 42.6 |
| 1 | 43.7 ± 54.2 | 58.2 ± 26.2 |
| 2 | 64.4 ± 135.1 | 58.3 ± 20.4 |
| 3 | 13.4 ± 7.9 | 51.1 ± 27.6 |
| 4 | 31.6 ± 26.9 | 76.4 ± 23.4 |
| Gene | Accession No. | Forward Primer | Reverse Primer |
|---|---|---|---|
| FXR | NM_001163700 | 5′-GGAGAGGAAGACTCAGTCCAGA-3′ | 5′-GCCTGGACGACAGAGCTAAA-3′ |
| BSEP | NM_021022 | 5′-GGACACACCTGAGAGGACCTT-3′ | 5′-AGATCGTTGACGGATGGAAG-3′ |
| SHP | NM_011850 | 5′-CTGAAGGGCACGATCCTCT-3′ | 5′-GCCTCCTGTTGCAGGTGT-3′ |
| ALP | NM_007431 | 5′-AACCCAGACACAAGCATTCC-3′ | 5′-GAGAGCGAAGGGTCAGTCAG-3′ |
| RUNX2 | NM_009820 | 5′-GCACCGACAGTCCCAACT-3′ | 5′-CTCCGAGGGCTACAACCTT -3′ |
| COL1A1 | NM_007742 | 5′-CATGTTCAGCTTTGTGGACCT-3′ | 5′-GCAGCTGACTTCAGGGATGT-3′ |
| OCN | NM_001032298 | 5′-AGACTCCGGCGCTACCTT-3′ | 5′-CTCGTCACAAGCAGGGTTAAG-3′ |
| GAPDH | NM_008084 | 5′-CAAGGAGTAAGAAACCCTGGACC-3′ | 5′-CGAGTTGGGATAGGGCCTCT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujimori, K.; Iguchi, Y.; Yamashita, Y.; Gohda, K.; Teno, N. Synthesis of Novel Farnesoid X Receptor Agonists and Validation of Their Efficacy in Activating Differentiation of Mouse Bone Marrow-Derived Mesenchymal Stem Cells into Osteoblasts. Molecules 2019, 24, 4155. https://doi.org/10.3390/molecules24224155
Fujimori K, Iguchi Y, Yamashita Y, Gohda K, Teno N. Synthesis of Novel Farnesoid X Receptor Agonists and Validation of Their Efficacy in Activating Differentiation of Mouse Bone Marrow-Derived Mesenchymal Stem Cells into Osteoblasts. Molecules. 2019; 24(22):4155. https://doi.org/10.3390/molecules24224155
Chicago/Turabian StyleFujimori, Ko, Yusuke Iguchi, Yukiko Yamashita, Keigo Gohda, and Naoki Teno. 2019. "Synthesis of Novel Farnesoid X Receptor Agonists and Validation of Their Efficacy in Activating Differentiation of Mouse Bone Marrow-Derived Mesenchymal Stem Cells into Osteoblasts" Molecules 24, no. 22: 4155. https://doi.org/10.3390/molecules24224155





